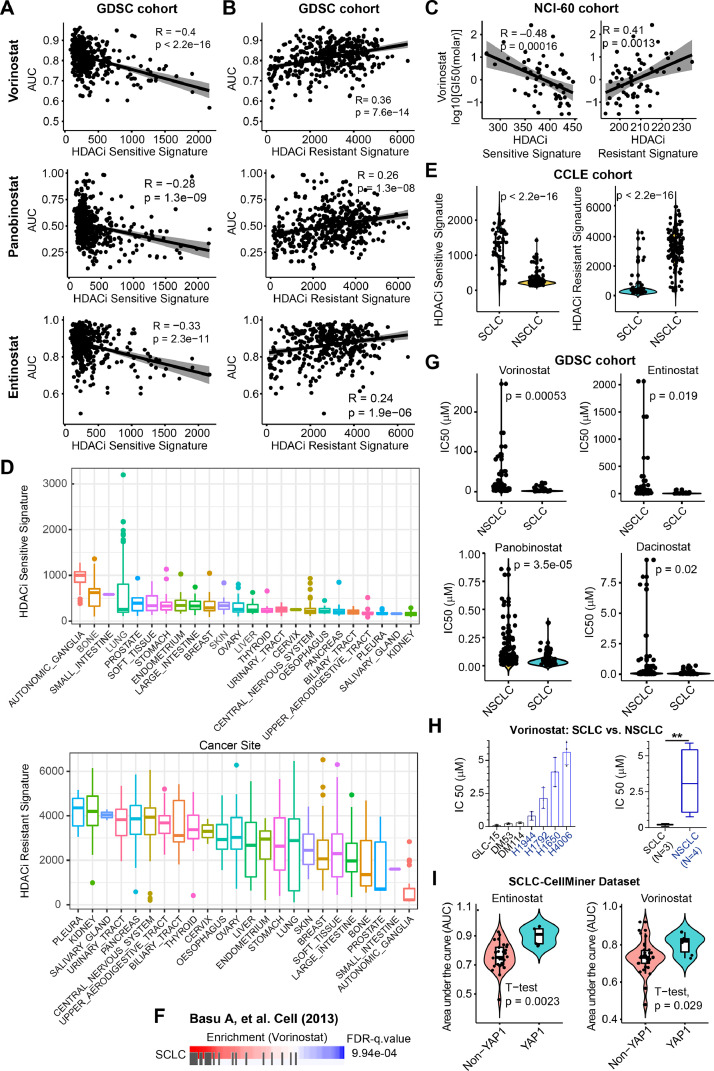
Fig. 2.
Generalized HDACis gene signatures reveal SCLC as potential subsets for therapeutic response to HDACis. A-B, An independent data cohort validating the correlation of HDACis sensitive (A) and resistant (B) signatures with the AUC (area under the curve) values of three HDACis (Vorinostat, Panobinostat, and Entinostat). Drug response profiles were downloaded from the GDSC (Genomics of Drug Sensitivity in Cancer; https://www.cancerrxgene.org/) database. C, An independent NCI-60 data cohort validating the correlation of HDACis sensitive (left) and resistant (right) signatures with –log10 [GI50(molar)] of Vorinostat. Drug response profiles were downloaded from CellMinerCBD (version 1.2) (https://discover.nci.nih.gov/cellminercdb/), and then merged with the gene expression array of the tested cancer cell lines. D, Prospective analysis of HDACis sensitive (upper panel) and resistant (lower panel) signature scores across CCLE (Cancer Cell Line Encyclopedia) pan-solid cancer lines. HDACis sensitive and resistant signature scores were calculated by summarizing the Z-normalized log2RSEM (RNA-Seq by Expectation-Maximization) of the expression data for the genes in the HDACis sensitive and resistant signatures, based on the RNA-sequencing data of pan-solid cancer cell lines from CCLE. E, Significant difference (by Welch's t-test) of HDACis sensitive (upper panel) and resistant (lower panel) signature scores between small cell lung cancer (SCLC) and NSCLC cells. F, Cancer cells with high sensitivity (in red) to HDACi Vorinostat are significantly enriched for SCLC. Data were from Basu A, et al. Cell (2013). G, Violin plots showing that SCLC cells are more sensitive to four HDACis reflected by the IC50 values, compared with NSCLC cells. H, Experimental validation of the differential sensitivity (reflected by IC50 value) of SCLC and NSCLC cells in response to HDACi Vorinostat. A significant difference was calculated by Welch's t-test. **p < 0.01 (by Welch's t-test). I, Independent datasets CellMiner-SCLC validating the association of non-YAP1-driven SCLC with therapeutic responses to two clinically-approved HDACis. Data were from a curated public dataset (https://discover.nci.nih.gov/SclcCellMinerCDB/).