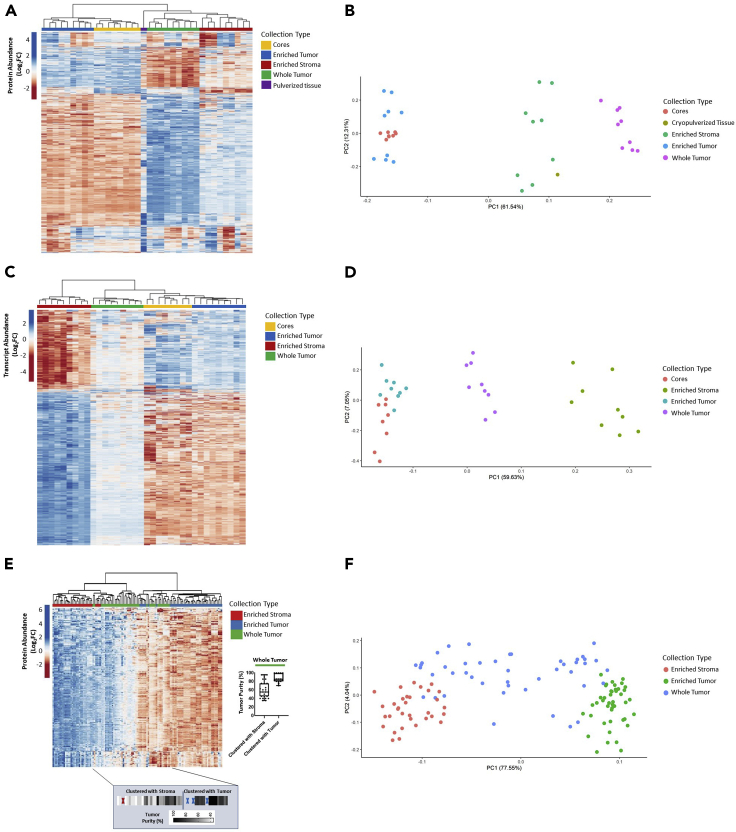
Figure 2.
Unsupervised hierarchical cluster analysis and principal component analysis of LMD-enriched samples by differentially abundant protein and transcript expression
(A, B) 1,928 differentially abundant proteins and (C, D) 3,861 transcripts with median absolute deviation (MAD) > 0.5 from case 343WM and (E, F) 6,199 differentially abundant proteins with MAD>1 in the 9-patient specimen set. (E, F) Protein abundances are represented across n = 123 samples derived from n = 9 patients consisting of LMD-enriched tumor epithelium (n = 45 total; 5 levels/patient), LMD-enriched stroma (n = 33 total; 2–5 levels/patient), and whole tumor (n = 45 total; 5 levels/patient) samples. Highlighted box depicts the median tumor purity values from manual pathology review for each of the whole tumor collections as they appear in the heatmap, ordered from left to right. Dark elongated border line distinguishes the whole tumor collections which clustered with LMD-enriched stroma (left) from those that clustered with LMD-enriched tumor (right). Red and blue “X” marks represent interceding LMD-enriched stroma or tumor collections, respectively.