Figure 2. RT-PCR and LAMP assays for detection of SARS-CoV-2 infection.
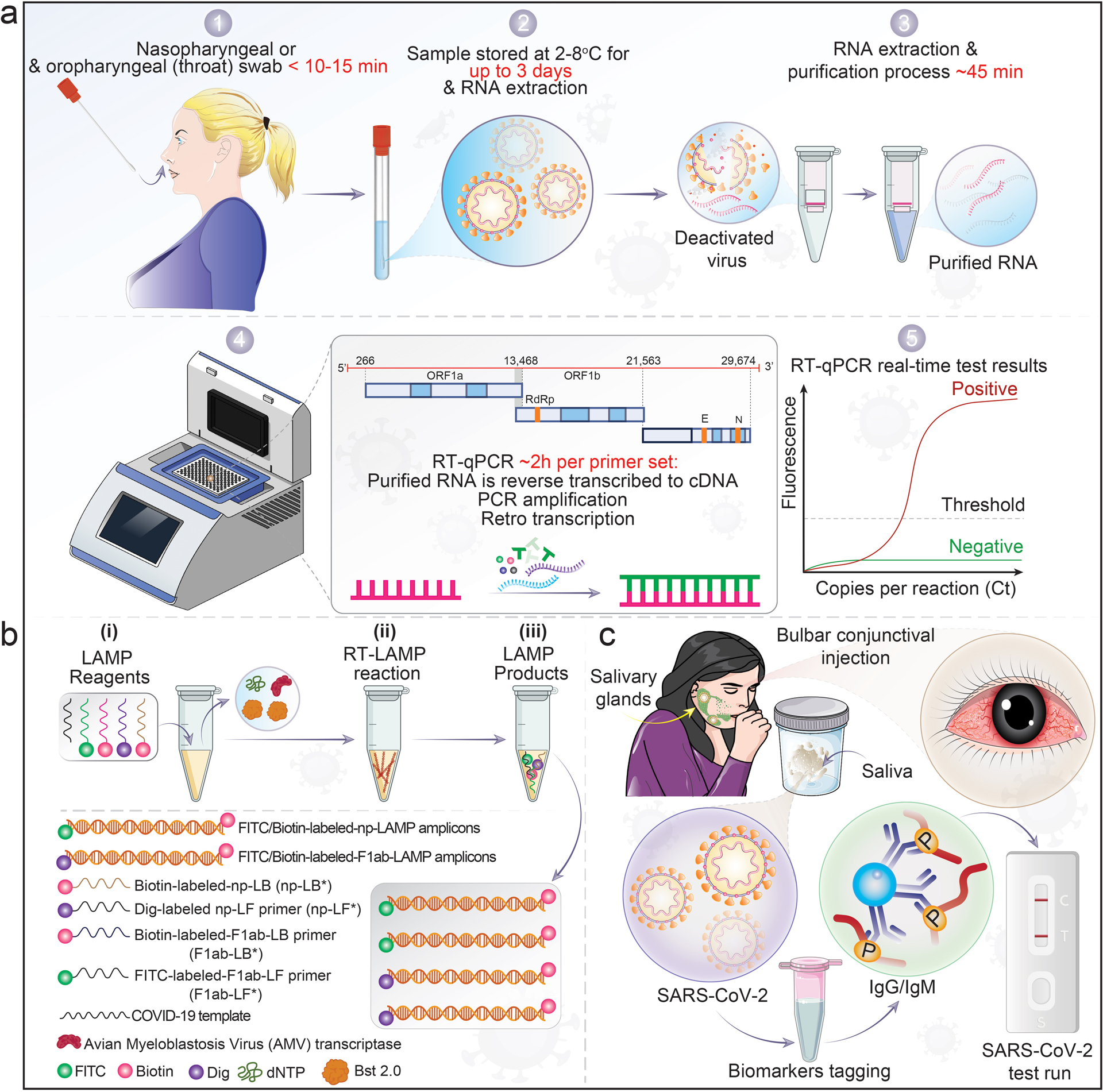
a, A nasopharyngeal swab collects patient samples. 1. RNA is extracted from fluids that contain SARS-CoV-2-infected cells and free viral particles, 2–3. The recovered viral RNA is then reverse transcribed to cDNA and amplified for detection of viral nucleic acids. 4. Conserved regions of the RdRP and E genes are the sub-genomic viral segments amplified with a fluorogenic probe by quantitative PCR. 5. Positive cases exceed the threshold of detection. b, Description of the SARS-CoV-2 RT-loop-mediated isothermal amplification (LAMP) assay. (i), amplification mixtures; (ii), RT-LAMP reaction, and (iii), the products of SARS-CoV-2 RT-LAMP reactions. Although RT-PCR methods are used as the standard for detection of SARS-CoV-2 due to high sensitivity, limitations are present. As an alternative, isothermal amplification methods or LAMP was developed. When optimized for detection, the assay is as sensitive as standard PCR detecting < 10 viral copies/reaction. c, SARS-CoV-2 saliva test. The illustration demonstrates SARS-CoV-2 infection in salivary glands and released specific biomarkers that accumulate in the oral cavity. These are collected through a sample tube and tagged with specific biomarker protein and run through lateral flow rapid tests.
