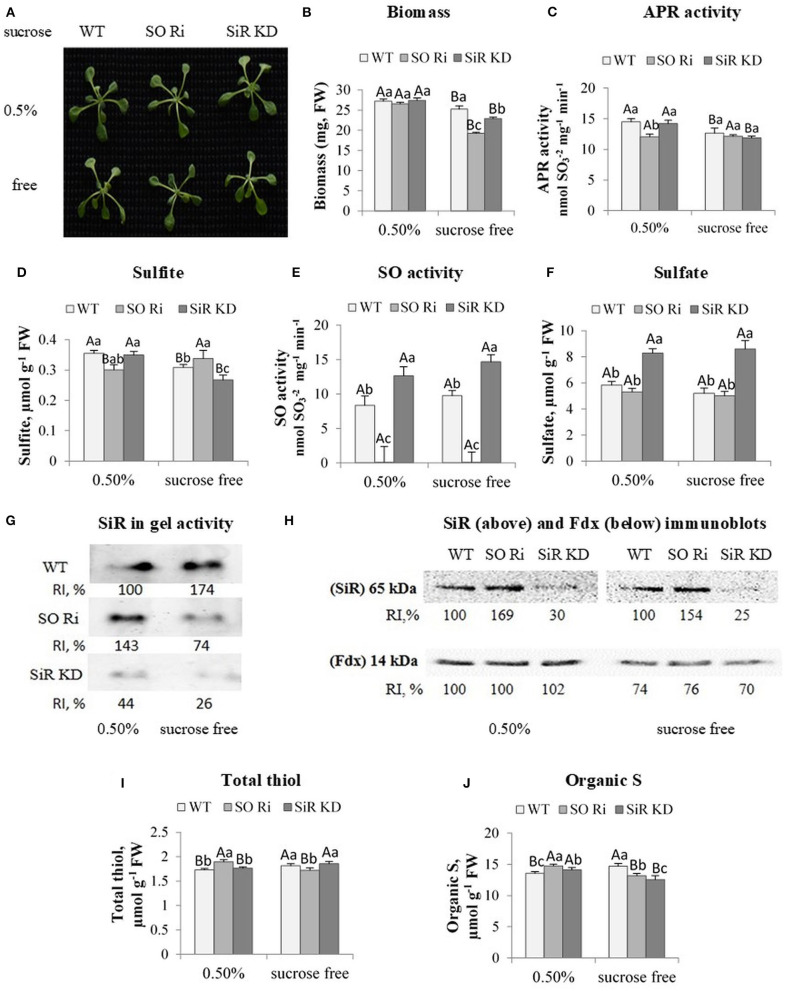
Figure 3.
Characterization of Arabidopsis wild-type (WT), as well as sulfite reductase (SiR KD) and sulfite oxidase (SO Ri), modified plants in response to 0.5% (normal) and 0% sucrose (sucrose-free) conditions. (A) Plants appearance, photographed 9 d after transfer, of 8-day-old seedlings, to agar plates containing 0.5 MS with or without sucrose. (B) Total biomass accumulation of the upper plants part. Values are means (n = 20 plants). (C) APR activity. (D) Sulfite level. (E) Activity of SO enzyme assayed as sulfite disappearance. (F) Sulfate level, (G). SiR in-gel activity indicated by the appearance of sulfide, the final product of SiR activity reacted with lead acetate to generate the brown precipitate of lead sulfide. Crude extracts of carbon treated plants were fractionation employing NATIVE PAGE conditions with two separated gels with control plants (see the full gels in Figure 4D). RI represents relative intensity (%) compared with the activity of WT treated with 0.5% sucrose as 100%. (H) Immunoblot analyses of Arabidopsis SiR (upper insert) and Fd (lower insert) proteins fractionated by SDS-PAGE and immunoblotted with SiR- and Fd-specific antiserums. Each lane contains 10 μg of proteins for SiR and 30 μg for Fd. Relative intensity, % (RI, %). (I) Total non-protein thiol and (J). Organic S level. The organic S was calculated as the difference between total S to the inorganic S (sulfate + sulfite). Values for (C–F,I,J) represent the means of three independent experiments (±SE, n = 3). (G,H) Represent one of three independent experiments with similar results. Different lowercase letters indicate differences between genotypes within the same treatment. Different uppercase letters indicate significant differences within the plant genotypes in response to treatment (Tukey–Kramer HSD test; JMP 8.0 software).