Figure 3. Dap can move from the oocyte to the NCs.
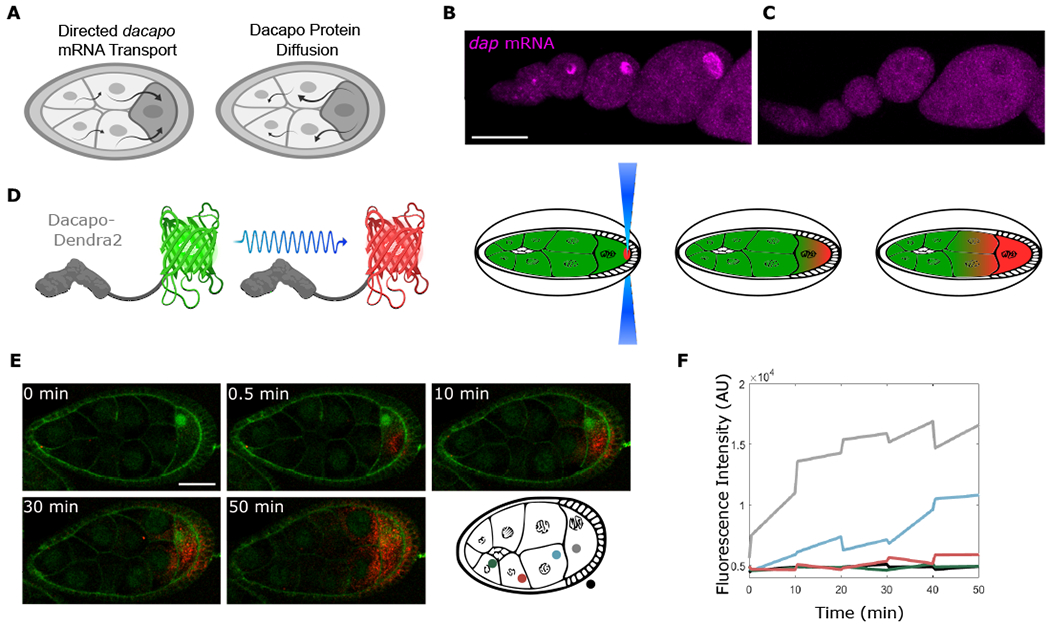
(A) Model predicts that the localized dap mRNA creates a pool of Dap protein that moves to the NCs.
(B and C) Single molecule fluorescence in situ hybridization (smFISH) for sfGFP mRNA performed on egg chambers from control Dap-sfGFP females (B) or from Dap-sfGFP females fed with yeast containing colchicine (C). ANOVA performed on fluorescence intensity collected from an ROI within the oocyte for stages 4-8 egg chambers, F value = 32.71, p value = 2.08 × 10−5; n = 10 egg chambers for each condition.
(D) Dap-Dendra2 photoversion scheme.
(E) Time sequence of Dap-Dendra2 photoconversion of region shown in (D). Photoconversion performed every 10 min. First photoconversion event (between t = 0 min and t = 0.5 min) was performed 3× to bleach Dendra2 in ROI, intended to mark region of excitation and confirm that nonspecific photoconversion does not occur in adjacent NCs. Successive photoconversion events were performed once. Membranes were stained with CellMask green plasma membrane stain.
(F) The graph shows fluorescence intensity quantified before and after each photoconversion event. Colors correspond to measurements in different regions of the egg chamber. Scale bars, 50 μm (B) and (C), 40 μm (E).
