Figure 1.
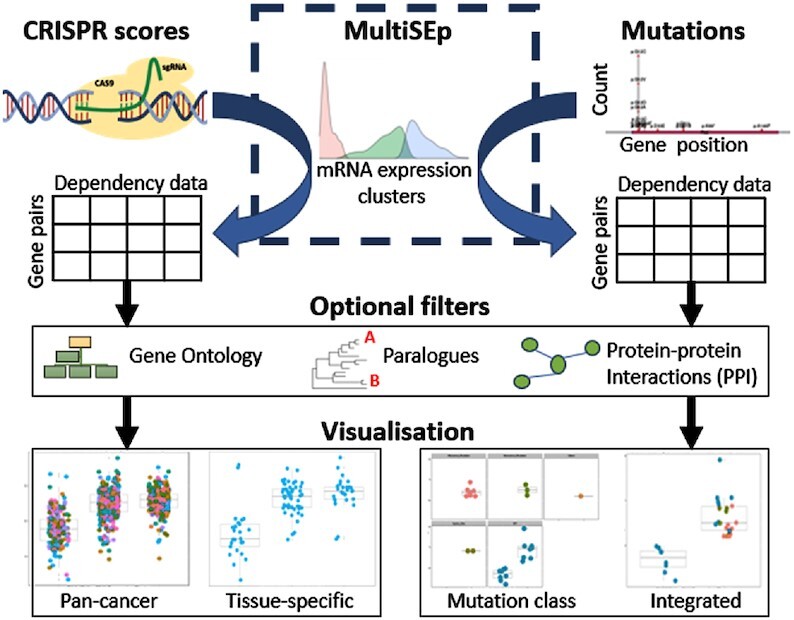
Overview of SynLeGG. CRISPR essentiality scores from CERES (12) and mutations from whole exome sequencing are analysed in separate workflows, partitioned using MultiSEp gene expression clusters. Results are returned as a table where each row describes a gene pair and the columns summarise dependency data, including q-values for the difference between CRISPR or mutation values across the MultiSEp clusters. Application of optional filters enables prioritization of gene pairs with orthogonal evidence of similar function according to common Gene Ontology terms, evolutionary information and protein interactions. Multiple visualizations and download of data are available to facilitate exploration of candidate gene dependency relationships.
