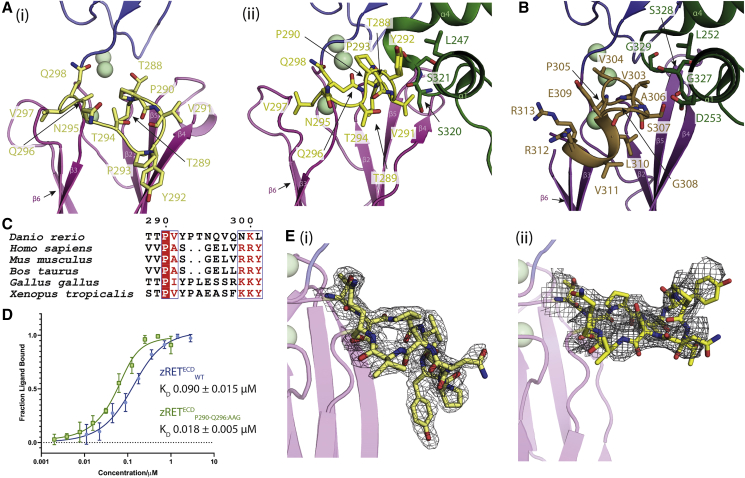
Figure 3.
Ligand-co-receptor-induced conformational changes in zRETECD
(A) The CLD3-β2-β3 loop is shown in yellow as sticks (i) projects “downward” in the view shown for zRET CLD(1-4) (see the orientation of Y292 side chain) and (ii) projects “upward” to engage the GFRα1D2 α1 helix (green sticks) in the zRGα1a structure.
(B) The shorter CLD3-β2-β3 loop and extra helix from the human RETECD-NRTN-GFRα2 structure (PDB: 6Q2O) shown as olive colored sticks, domains colored as in Figure 1.
(C) Sequence alignment of RET CLD3-β2-β3 loop sequences by Espript (http://espript.ibcp.fr) (Robert and Gouet, 2014).
(D) Binding curves and calculated KD values for zRETECDwt and mutant (zRETECDP291-Q296:AAG) binding to zGFRα1a2-zGDNF2 measured by MST, with a minimum of n = 3 repeats for the WT and the mutations with the SE represented.
(E) (i) Electron density map calculated using m2Fo-DFc coefficients over the CLD3-β2-β3 loop, yellow sticks and contoured at 1.0σ. (ii) Coulombic potential cryo-EM map for CLD3-β2-β3 loop from the zRGα1a complex (black mesh). Calcium ions are represented as pale green spheres.