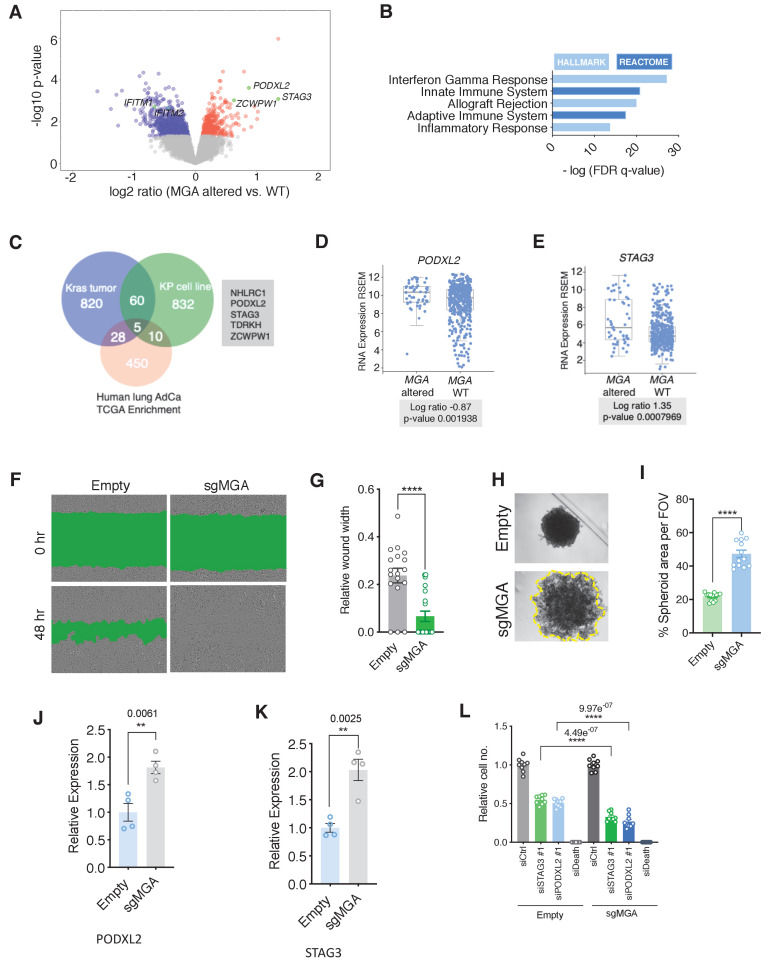
Figure 7. MGA loss correlates with activation of STAG3 and PODXL2 in human lung cancer and results in a pro-invasive phenotype in vitro.
(A) Volcano plot of transcripts that are differentially expressed in MGA altered (n = 52) vs. WT (n = 455) from pancancer TCGA lung adenocarcinoma data. Representative genes highlighted in green. (B) Hallmark and Reactome analysis of genes downregulated in MGA altered patients vs. non-altered (p-value cutoff < 0.05). (C) Venn diagram depicting overlap between genes upregulated in the mouse Kras tumor, KP cell line, and Human lung TCGA data with MGA alterations. (D, E) Log ratios of (D) PODXL2 and (E) STAG3 expression in MGA altered vs. WT lung adenocarcinoma patients (TCGA, 2014). (F) Representative wound widths and (G) quantification of wound width at 48 hr in Empty and sgMGA wells (n = 18 Empty, n = 20 sgMGA. ****p=4.77e−05). (H) Spheroid formation and (I) quantification in Empty and sgMGA A549 lines (quantification at Day 6, n = 13 spheroids for each from three independent experiments. ****p=4.89e−11). (J, K) qPCR for (J) PODXL2 and (K) STAG3 expression in Empty and sgMGA A549 cells (n = 4 replicates for each condition). (L) Cell growth upon siRNA knockdown of STAG3 and PODXL2 in A549 Empty and sgMGA cells (n = 3 replicates from three independent experiments for each group). All p-values calculated using a two-sided Student’s t-test unless otherwise noted. Error bars represent SEM.