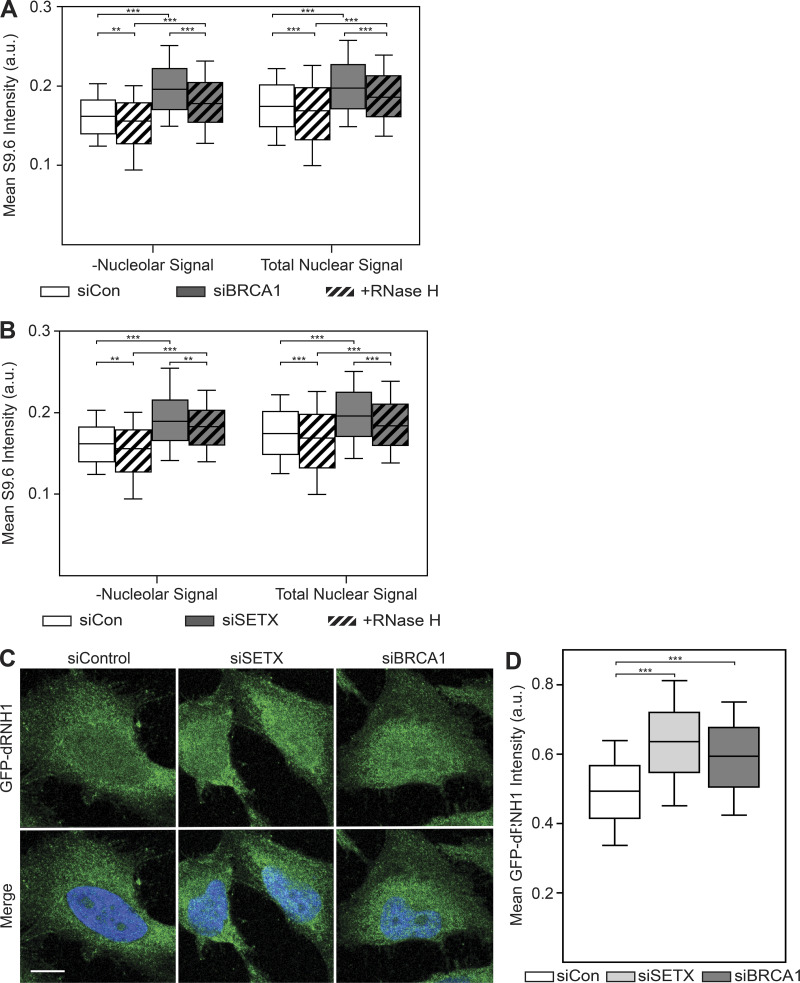
Figure S4.
Exclusion of nucleolar regions does not resensitize S9.6 immunostaining to RNase H treatment, whereas GFP-dRNH1 is compatible with PFA fixation.(A) Quantification of mean nuclear S9.6 signal in control or siBRCA1 cells after excluding the nucleolar regions (left). Total mean nuclear S9.6 signal after BRCA1 depletion (Fig. 3 B) is shown on the right for reference. (B) Same as in A but with SETX depletion. Nucleolar regions were excluded (left), whereas total mean nuclear S9.6 signal after SETX depletion (Fig. 3 E) is shown on the right for reference. (C) Representative confocal images of 4% PFA-fixed HeLa cells. After fixation, coverslips were incubated with GFP-dRNH1. GFP-dRNH1 signal is shown in green and DAPI in blue. (D) Quantification of mean nuclear GFP-dRNH1 intensities for the conditions shown in C. Box plots show median (box central line), 25% and 75% percentiles (box edges), and 10% and 90% percentiles (whiskers). Data are combined from three biological replicates (n = 3), with at least 100 nuclei scored per condition per experiment. **, P ≤ 0.01; ***, P ≤ 0.001 (Mann-Whitney U test). Scale bars are 10 microns.