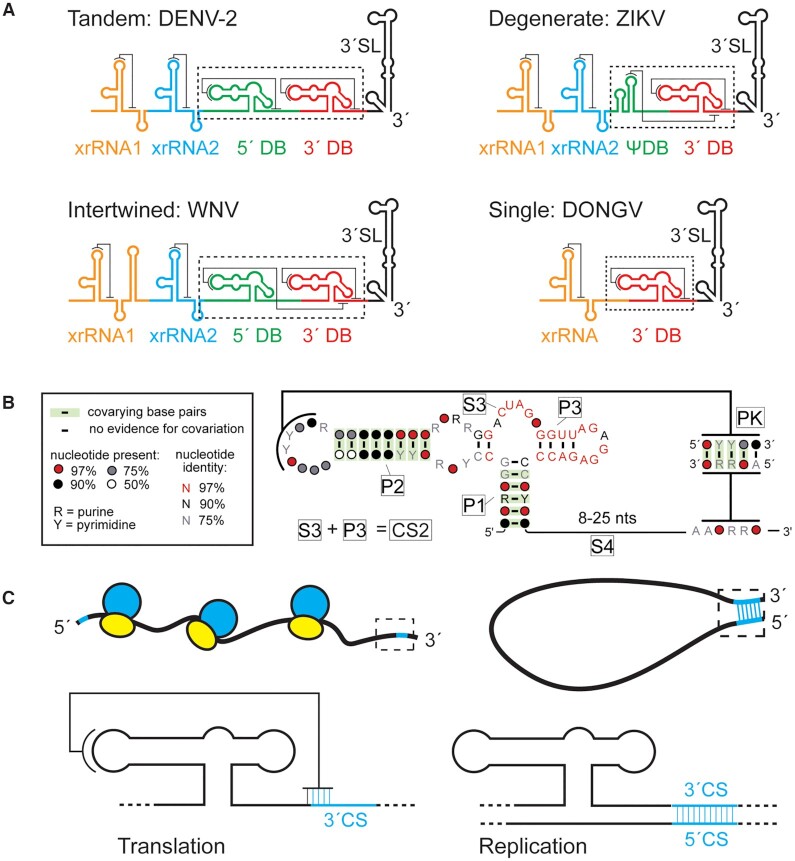
Figure 1.
Conservation and role of flaviviral DB RNAs. (A) Predicted organization and predicted secondary structures of 3′ UTRs from DENV-2, West Nile virus, ZIKV, and DONGV. The 3′ dumbbell element is conserved but can exist in several configurations. (B) Co-variation analysis of the 3′ DB element from 46 different viruses analyzed using R-scape (40). Predicted helices (P1, P2, P3 and PK) are labeled as are predicted unpaired linker regions (S3, S4). The S3 and P3 regions collectively make up a previously identified conserved sequence (CS2) (3). (C) Model of the 3′ DB element during translation and replication. The pseudoknot overlaps with the 3′ CS, an element responsible for base pairing with the 5′ CS during viral minus-strand synthesis. During viral protein translation (left) the DB pseudoknot is formed. The pseudoknot is incompatible with 5′-3′ CS formation (cyan) during replication (right).