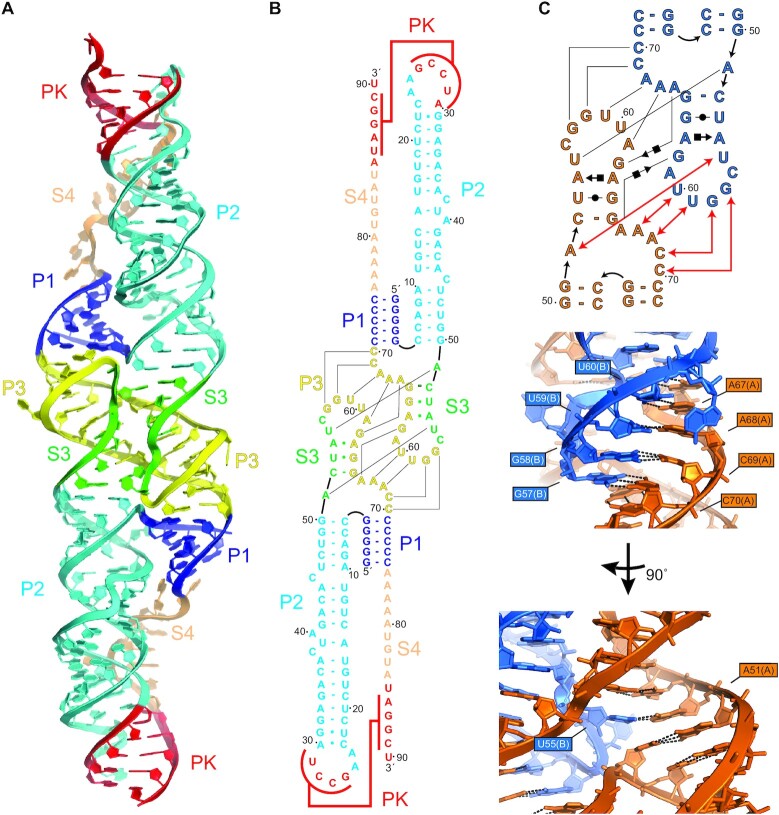
Figure 2.
Crystal structure of the DONGV DB RNA. (A) Overall structure of the dimer form of the wild-type DONGV DB observed in the crystal. Bound iridium (III) hexammine ions used for SAD phasing are omitted for clarity. Predicted stems from Figure 1B are labeled (P1 = blue, P2 = cyan, P3 = yellow, pseudoknot = red) as are the predicted unpaired regions (S3 = green, S4 = orange). (B) Base pairing interactions observed in the RNA crystal structure colored as in (A). Lines indicate base pairing observed between copy 1 and copy 2. (C) Secondary structure in Leontis-Westhof notation (top) and detailed 3D view (bottom) of the dimer interface highlighting in trans base pairing interactions between molecule A (orange) and molecule B (blue). Red lines in the secondary structure show base pairing interactions labeled in the 3D view.