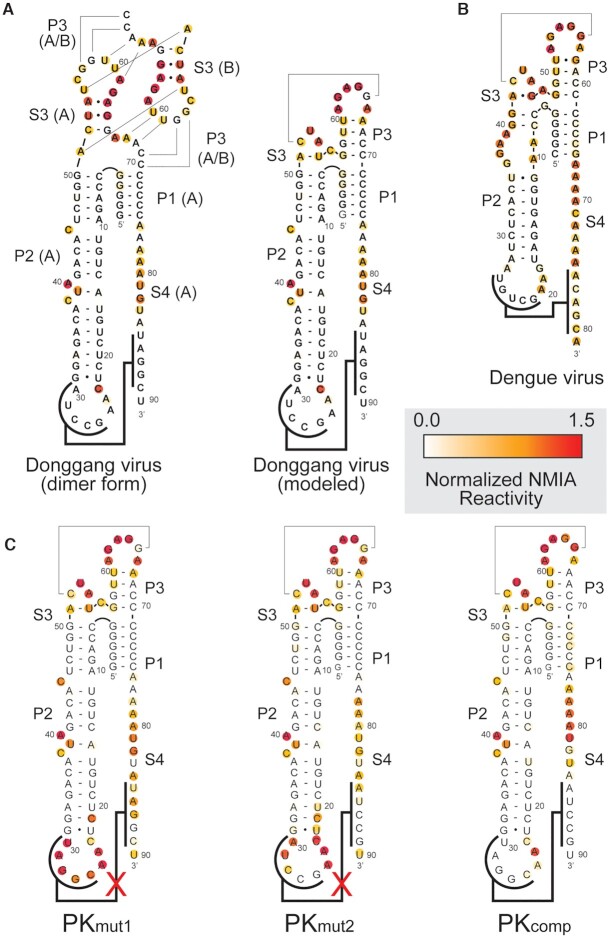
Figure 5.
NMIA chemical probing of the DB RNA. (A) Secondary structure of the DONGV DB RNA in the form observed in the crystal structure (dimer form) compared with the modeled form. NMIA reactivity of the wild-type DB sequence is color-coded (red is high reactivity, white is low reactivity). (B) NMIA reactivity of the DENV-2 DB plotted on the putative secondary structure based on homology to DONGV. (C) NMIA reactivity of pseudoknot mutant RNAs (PKmut1 and PKmut2) plotted on the modeled RNA secondary structure. The mutations in PKmut1 and PKmut2 were combined to generate compensatory base pairing (PKcomp).