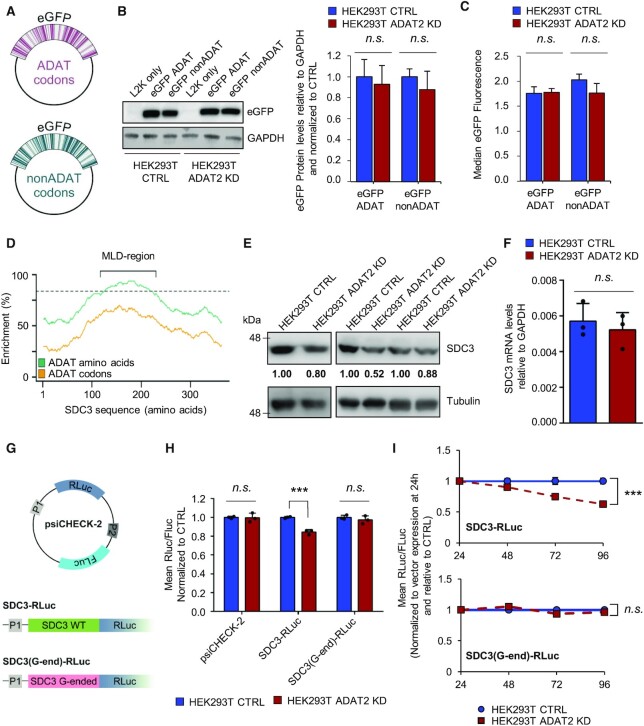
Figure 7.
(A) Schematic representation of constructs encoding eGFP with TAPSLIVR codons that are C-ended (ADAT codons, upper panel in violet) or G-ended (nonADAT codons, lower panel in dark green). (B) eGFP protein levels in the indicated cell lines when transfected with eGFP constructs depicted in (A). GAPDH is used as gel loading control. ‘L2K only’: cells incubated with lipofectamine 2000 in the absence of eGFP constructs. Quantification of western blot bands for eGFP relative to GAPDH levels and normalized to CTRL cells was obtained from three biological replicates. Shown are the mean and SD. n.s.: not statistically significant (t-test). See also Supplementary Figure S5A–C. (C) Evaluation of eGFP fluorescence by FACS in the indicated cell lines when transfected with eGFP constructs depicted in (A). Shown is the median and SD of biological duplicates. n.s.: not statistically significant (t-test). (D) Distribution of TAPSLIVR (ADAT amino acids; green curve) and of codons decoded by I34-tRNAs (ADAT codons; orange curve) along the coding sequence of SDC3. Dotted line indicates the threshold of TAPSLIVR enrichment considered significant as defined in (23). The low-complexity TAPSLIVR-rich region containing the Mucin-like domain (MLD) is shown. (E) SDC3 protein levels in HEK293T CTRL and ADAT2 KD cells. Tubulin is used as gel loading control. Quantification of SDC3 bands relative to Tubulin and normalized to CTRL cells for each biological triplicate is shown. (F) SDC3 transcript levels relative to GAPDH in the indicated cell lines. Shown are biological triplicates, their mean and SD. n.s.: not statistically significant (t-test). (G) Schematic representation of psiCHECK-2-based SDC3 luciferase reporters bearing the wild-type low-complexity region of SDC3 (‘SDC3 WT’ in light green) or the same amino acid sequence but encoded with synonymous G-ended codons for all TAPSLIVR (codons not recognised by I34-tRNAs, ‘SDC3 G-ended’ in pink). The promoter 1 (P1) drives the expression of Renilla luciferase (RLuc). The second independent promoter (P2) drives the expression of Firefly luciferase (FLuc). (H) Evaluation of luciferase expression (RLuc/FLuc ratio normalized to CTRL cells) in HEK293T CTRL and ADAT2 KD cells transfected with the constructs depicted in (G). Shown are biological triplicates, their mean and SD. n.s.: not statistically significant. **: P-value < 0.01 (t-test). (I) Time-course analysis of luciferase expression (RLuc/FLuc ratio normalized to CTRL cells and to vector expression at the 24 h time-point) in the indicated cell lines transfected with SDC3-RLuc (upper panel) or SDC3(G-end)-RLuc (lower panel). Shown are the means (blue for CTRL and red for ADAT2 KD cells) and SD of biological triplicates. n.s.: not statistically significant. ***: P-value < 0.001 (t-test). See also Supplementary Figure S5D.