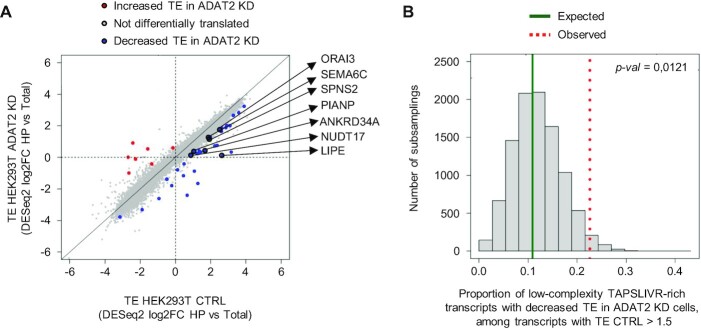
Figure 8.
(A) Scatter plot showing the evaluation of translation efficiency (TE) for individual transcripts based on an interaction analysis between transcript expression in total RNA (Total) and high polysome (HP) fractions obtained for experiments as in Figure 6. Blue, Red and Grey dots indicate genes with decreased TE, increased TE and unchanged TE in HEK293T ADAT2 KD cells as compared to HEK293T CTRL cells, respectively. Note that the x- and y-axis show Log2FC values (log2FC 1.5 = 0.58). Translationally impaired transcripts that encode proteins with low-complexity TAPSLIVR-rich regions are indicated. See also Supplementary Table S4. (B) Histogram of a permutation test using 10 000 sets of 31 random transcripts detected in experiments shown in (A). The expected (dark solid green line) and observed (red dotted line) proportion of transcripts encoding low-complexity TAPSLIVR-rich proteins with decreased TE in ADAT2 KD cells among transcripts highly translated in HEK293T CTRL cells (FC HP CTRL versus Total CTRL > 1.5) is indicated. P-values are computed as the proportion of permutations with more extreme statistics than the observed. See also Supplementary Table S4.