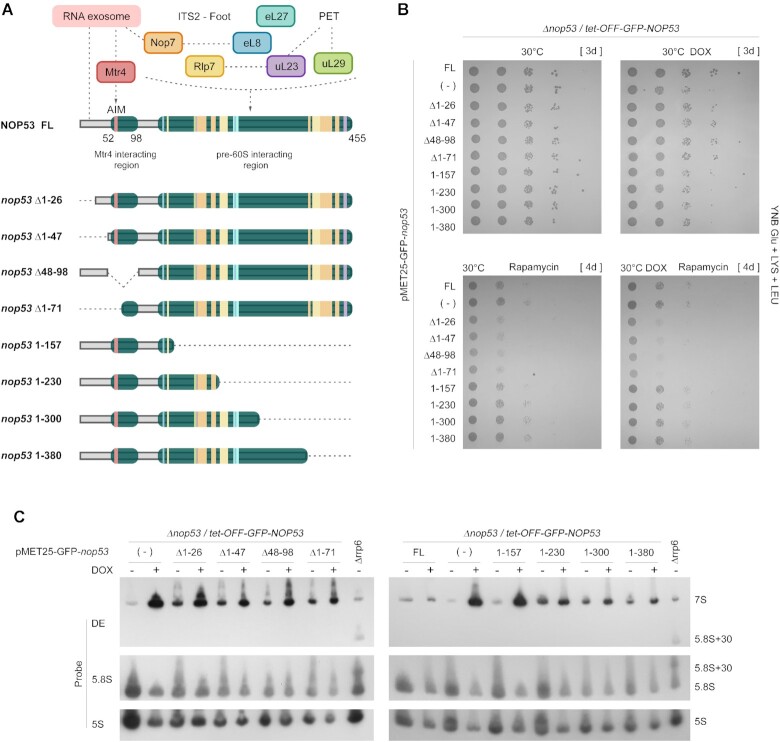
Figure 6.
Effect of nop53 truncation mutants on cell growth and pre-rRNA processing. (A) Schematic representation of the nop53 N- and C-terminal truncated mutants (Δ1–26, Δ1–47, Δ48–98, Δ1–71, 1–157, 1–230, 1–300, 1–380) indicating the regions involved in protein-protein interactions. The exosome and the RNA helicase Mtr4 interact with Nop53 N-terminus where the conserved AIM is found. 60S AFs (Rlp7, Nop7) and r-proteins (eL8, uL23, eL27, and uL29) which are associated with the ITS2-containing foot or PET directly interact with Nop53 at different points along the middle region and the C-terminus. (B) Spot growth assays performed with the conditional strain Δnop53/tet-OFF-GFP-NOP53, expressing the positive control GFP-Nop53 full length (FL), the negative control empty vector (−), or the indicated GFP-tagged nop53 N- and C-terminal truncated mutants. These strains were spotted in 10-fold serial dilution on to YNB Glu +LYS+LEU solid medium supplemented or not with doxycycline (DOX – 1.5μg/ml) and rapamycin (400 nM). They were incubated at 30°C for the period indicated in square brackets. (C) Northern blot analysis of total RNA extracted from the same strains grown at 30°C with or without doxycycline (DOX – 1.5 μg/ml) to evaluate the effect of the nop53 truncated mutants on the pre-rRNA processing. The RNA resolved through 8% polyacrylamide denaturing gel electrophoresis was analyzed using the indicated probes. The Δrrp6 strain was employed as a control for the accumulation of 5.8S + 30 pre-rRNA. All tested nop53 truncated mutants accumulated 7S pre-rRNA, although at different levels. The quantification of the bands is shown in Supplementary Figure S8D.