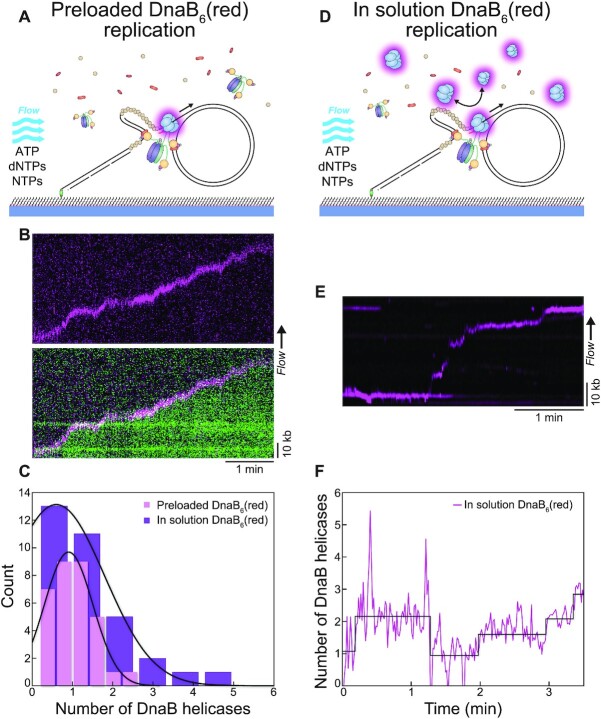
Figure 3.
More than one DnaB helicase are frequently present at the replication fork. (A) Illustration of the rolling-circle replication assay with DnaB6(red) preloaded on the DNA prior to introducing the replication solution. Replication is monitored in real time by flow stretching the replicating DNA products by hydrodynamic force. (B) (Top) Representative kymograph of preloaded DnaB6(red) moving with the fork during rolling-circle replication. (Bottom) Overlay of the DnaB6(red) and SYTOX Orange-stained DNA (green) kymographs shows the helicase molecule moving with the replication fork at the tip of the DNA product. (C) Distributions of DnaB helicase stoichiometry at the fork in the absence of DnaB6(red) in solution (magenta; n = 32) and in the presence of DnaB6(red) in solution (purple; n = 33). The black lines represent Gaussian fits to the data. (D) Illustration of the in-solution assay, where DnaB6(red) is preloaded and also included at 2 nM in the replication solution. (E) Representative kymograph showing the DnaB6(red) signal at the fork when DnaB6(red) is present in solution. (F) Number of DnaB6(red) as a function of time for the kymograph in (E) showing the fluctuation in DnaB helicase stoichiometry during the course of replication, where steps are detected by change-point analysis (47–49).