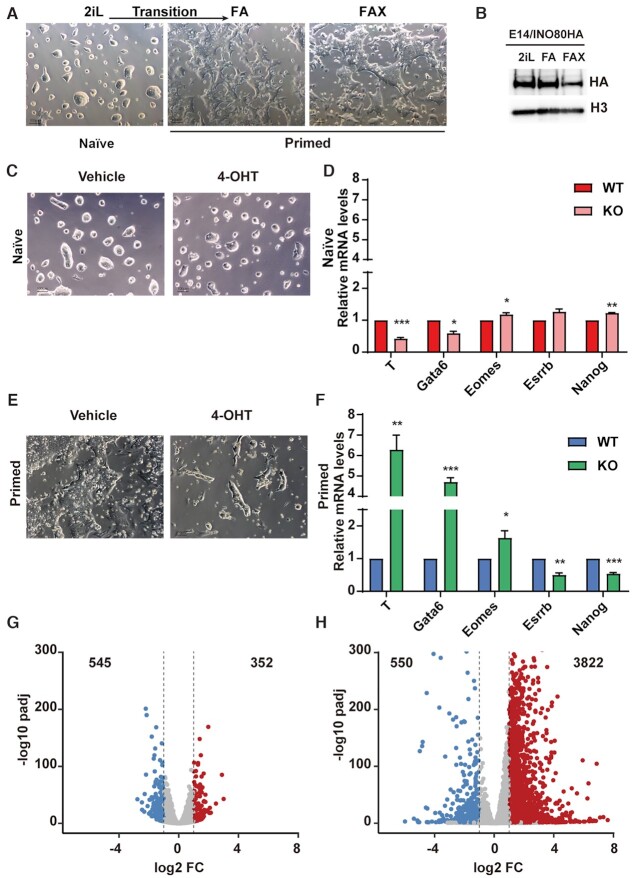
Figure 1.
INO80 is selectively required in the primed pluripotent state. (A) Culture model for the naïve and primed pluripotent state. Mouse ESCs were cultured in 2iL for the naïve state, switched to FA to induce the transition toward the primed state, and maintained in FAX for the primed state. Scale bar = 100 μm. (B) INO80 expression in different pluripotent states. Ino80-HA knock-in ESCs were cultured in 2iL, FA, or FAX, and INO80 expression was detected by western blot using the HA antibody. Histone H3 was blotted as the loading control. The experiment was repeated three times and one representative result was shown. (C, D) Ino80 deletion in the naïve state. Ino80-cKO ESCs were cultured in 2iL and treated with DMSO or 4-OHT for 2 days to induce Ino80 deletion. Cell morphology (C) and lineage marker expression (D) were examined by imaging and RT-qPCR. Scale bar = 100 μm. Relative mRNA levels between wild-type and Ino80 KO cells were first normalized by Gapdh and then normalized to wild-type cells. Results from three independent experiments were plotted as mean ± SEM. p-values were calculated by Student's t-test: * <0.05, ** <0.01, *** <0.001. (E, F) Ino80 deletion in the primed state. Ino80-cKO ESCs were cultured in FAX and treated with DMSO or 4-OHT for 2 days. Cell morphology (E) and lineage marker expression (F) were examined by imaging and RT-qPCR. Results from three independent experiments were plotted as mean (G) ± SEM. p-values (H) were calculated by Student's t-test: * <0.05, ** <0.01, *** <0.001. (G, H) Gene expression changes after Ino80 deletion in the naïve and primed state. Differentially expressed genes (DEGs) were determined by RNA-seq from two replicate experiments and highlighted in the volcano plots. Numbers of DEGs were also listed. Blue: down-regulated genes; Red: up-regulated genes.