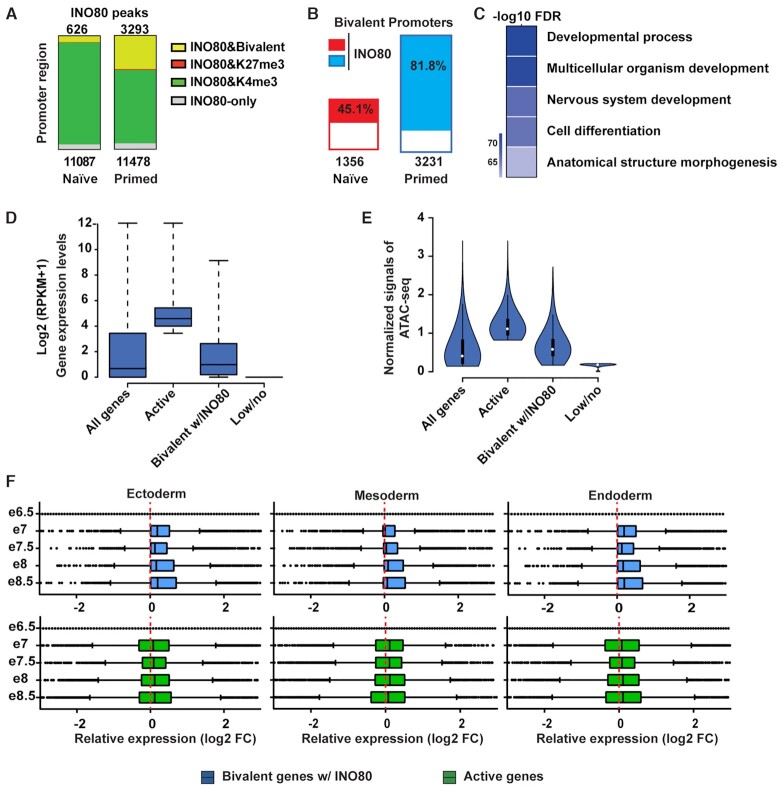
Figure 2.
INO80 occupies bivalent gene promoters. (A) H3K4me3, H3K27me3 and bivalent domains in INO80 occupied promoters in the naïve and primed state. Number of total INO80 peaks (bottom) and INO80 peaks with bivalent marks (top) in the promoter region were listed. (B) Percentage of bivalent promoters occupied by INO80 in the naïve and primed state. Number of total bivalent promoters were listed at the bottom. (C) Gene Ontology analysis of INO80 bound bivalent genes in primed state. Heatmap shows the Author: –log10(FDR) values of selected top enriched categories. The complete list of enriched categories can be found in Supplementary Figure S2I. (D) Box plot to show the relative expression of all genes, active genes (top 25%), INO80-bound bivalent genes, and lowly or not-expressed genes (bottom 25%) in the primed state. Gene expression was determined by RNA-seq and calculated as log2(RPKM + 1). (E) Violin plot of normalized ATAC-seq signals to show the chromatin accessibility of all genes, active genes (top 25%), INO80-bound bivalent genes, and lowly or not-expressed genes (bottom 25%) in the primed state. (F) Box plots to show relative gene expression changes of active and INO80-bound bivalent genes as in D during mouse embryonic development from E6.5-E8.5. Gene expression changes between Ectoderm, Mesoderm, or Endoderm cells and E6.5 epiblast cells were calculated as log2(RPKM + 1) from published single cell RNA-seq data (E-MTAB-6967).