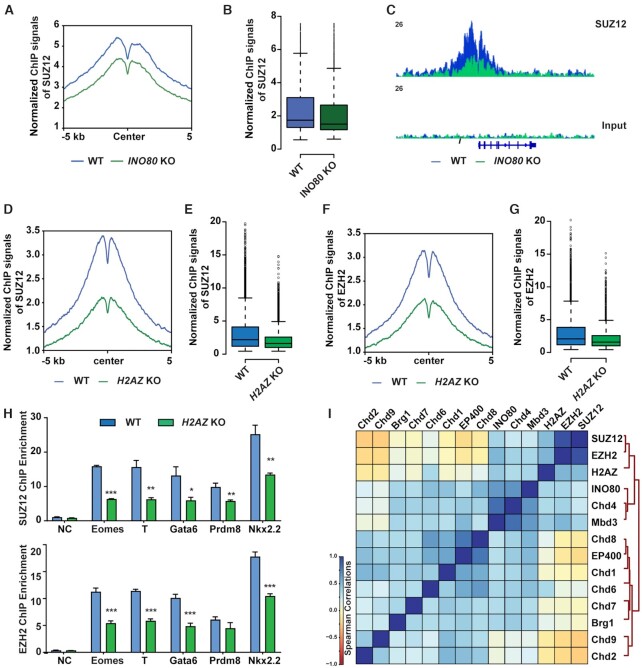
Figure 7.
INO80-dependent H2A.Z occupancy is required for PRC2 recruitment. (A, B) SUZ12 occupancy at INO80-bound bivalent TSSs in the primed state in WT and Ino80 deletion cells. Ino80-cKO ESCs were cultured in FAX and treated with 4-OHT for 2 days. Cells were cultured for 2 additional days and collected for ChIP-seq. SUZ12 occupancy was shown by metagene (A) and box (B) plots using normalized ChIP-seq signals. (C) Genome browser track to show SUZ12 occupancy near T in WT and Ino80 deletion cells. (D–G) SUZ12 and EZH2 occupancy at INO80-bound bivalent TSSs in the primed state in WT and H2az1/H2az2 deletion cells. H2az1-cKO/H2az2-KO ESCs were cultured in the primed state in FAX and treated with DMSO or 4-OHT for 2 days. Cells were cultured for another 2 days and collected for ChIP-seq. SUZ12 and EZH2 occupancy was shown by metagene (D, F) and box (E, G) plots using normalized ChIP-seq signals. (H) ChIP-qPCR to show SUZ12 (upper) and EZH2 (lower) at representative bivalent gene promoters in WT and Ino80 deletion cells. Fold enrichment was plotted as mean ± SEM from three replicates and p-values were calculated by Student's t-test: * <0.05, ** <0.01, *** <0.001. (I) Spearman correlation of genomic occupancy among chromatin factors. Except for that of INO80 and H2AZ, genomic occupancy of all other chromatin factors was based on public datasets (GSE64825).