Figure 1. Adenine base editing converts sickle cell disease β-globin (HBBS) to benign Makassar β-globin (HBBG) in patient CD34+ HSPCs.
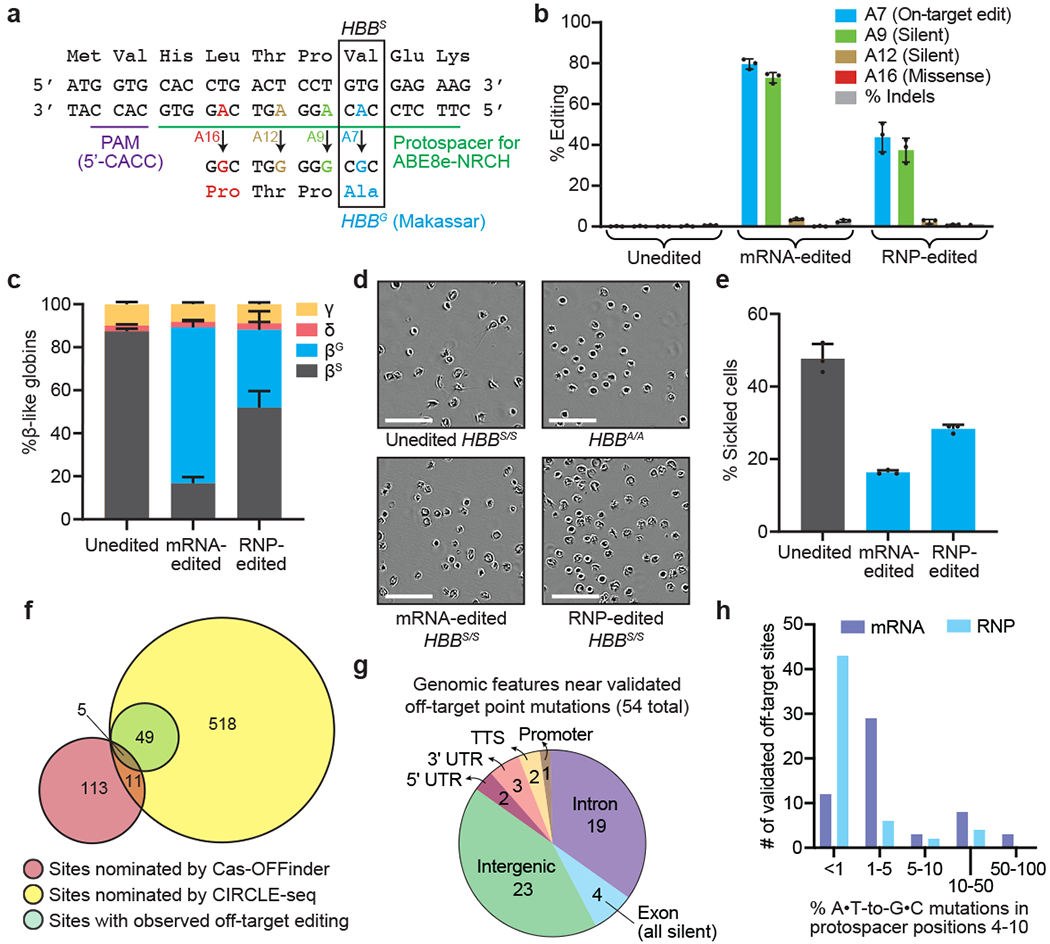
CD34+ cells from three SCD patient donors were electroporated with ABE8e-NRCH mRNA or RNP using an sgRNA targeting the SCD mutant HBB codon. (a) The edited region of HBB with the target A at protospacer position 7 shown in blue along with potential bystander edits in green (silent), brown (silent), and red (non-silent). (b) Editing efficiencies by HTS at target and bystander adenines, and indels after 6 days in stem-cell culture media following electroporation. (c) Proportion of β-like globin proteins by HPLC of reticulocyte lysates after 18 days in differentiation media following electroporation. (d) Representative phase-contrast images of reticulocytes derived from unedited or edited donor HSPCs incubated 8 hours in 2% O2. Nine images of >50 cells each were collected per sample. Scale bar=50 μm. (e) Quantification of sickled reticulocytes from counting >300 randomly selected cells by a blinded observer from images as in (d). (f) Venn diagram showing candidate off-target sites nominated by Cas-OFFinder and CIRCLE-seq, and nominated sites for which off-target editing was observed by targeted DNA sequencing in SCD patient CD34+ cells electroporated with ABE8e-NRCH mRNA. (g) Predicted genomic features of validated off-target sites. TTS, ≤1 kb from the transcription termination site; UTR, untranslated region. (h) ABE8e-NRCH-treated HSPCs from two different SCD patient donors were sequenced at 697 potential off-target sites. The histogram shows the number of validated off-target base editing sites binned by average percentage of sequencing reads for each site with any A•T-to-G•C mutations in protospacer nucleotides 4-10. Bar values in (b), (c), and (e) and error bars reflect mean±SD of three independent biological replicates, with individual values shown as dots.
