Extended Data Figure 4. Off-target base editing associated with ABE8e-NRCH conversion of HBBS to HBBG Makassar in sickle cell disease patient CD34+ hematopoietic stem and progenitor cells.
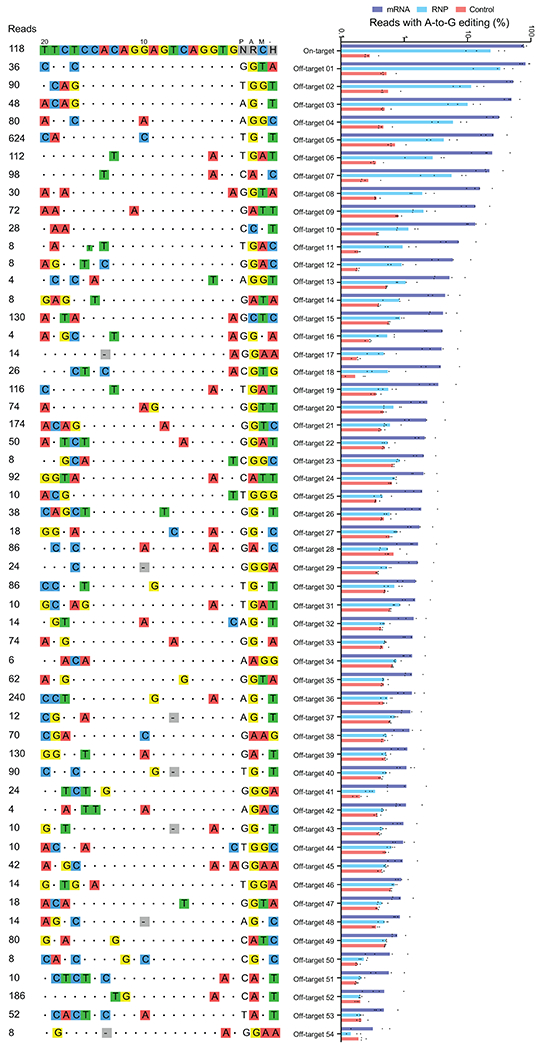
CIRCLE-seq read counts obtained for each verified off-target site and the alignment of each site to the guide sequence are shown. Bar graphs show the percentage of sequencing reads containing A•T-to-G•C mutations within protospacer positions 4-10 at on-and off-target sites in genomic DNA samples from patient CD34+ HSPCs treated with ABE8e-NRCH mRNA, protein, or untreated controls (n=4). Note that the mutation frequency shown is summed across all reads with one or more A•T-to-G•C mutations in this window. Sequencing errors therefore accumulate in control samples compared to standard sequencing error frequencies for a single nucleotide. Bar values and error bars reflect mean±SD.
