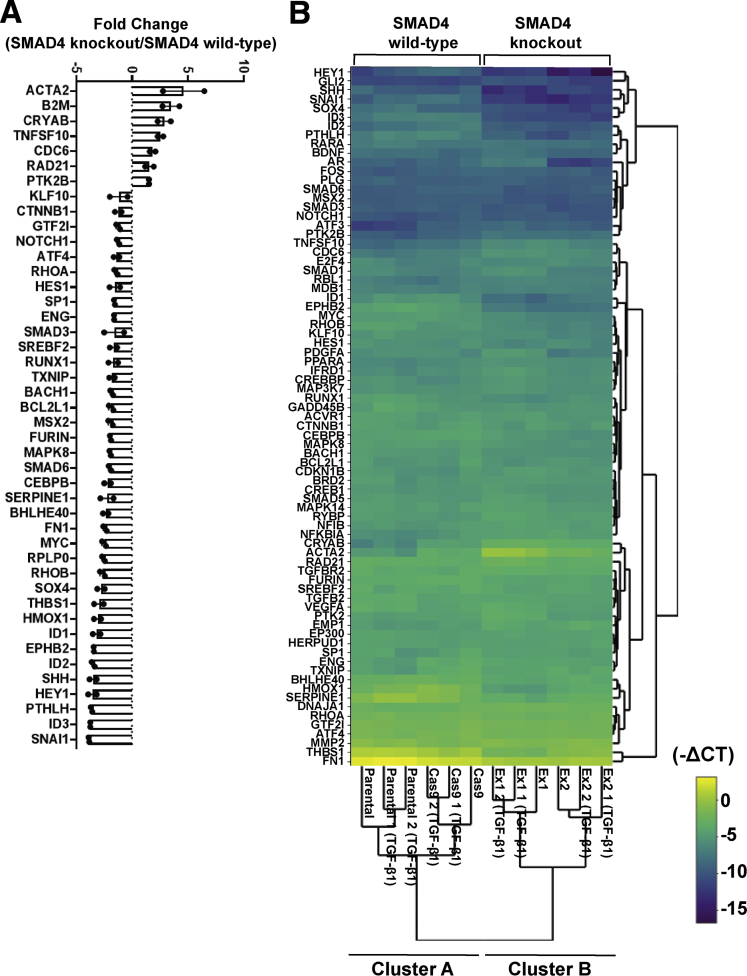
Figure 3.
Differential expression of TGF-β signaling target genes in SMAD4 knockout compared to SMAD4 wild-type cells. (A) Relative expression levels of TGF-β signaling target genes (RT2 Profiler PCR Array) in SMAD4 knockout cells (CP-B Ex1 and CP-B Ex2) vs SMAD4 wild-type cells (CP-B Cas9 only) under basal conditions (serum deprivation) for 16 hours. Thresholds for fold change ≥1.5 and ≤0.7 are chosen for up-regulated and down-regulated genes, respectively. Reciprocal fold change values are shown (y=0 or fold change 1 (no change) and y=-2 or fold change 0.5). N=1 experiment per each cell line (SMAD4 wild-type = CP-B Cas9 only cells; SMAD4 knockout = Ex1 and Ex2 clones). The comparison was made between CP-B Cas9/Ex1 and CP-B Cas9/Ex2 and mean fold change ± SEM is shown. No error bars indicate SEM less than the symbol size. (B) Hierarchical clustering indicating differentially expressed genes across individual samples, including 2 independent replicates per each of CP-B parental and Cas9 (SMAD4 wild-type), and Ex1 and Ex2 (SMAD4 knockout) cells following treatment with 10ng/mL TGF-β1 in serum deprivation for 16 hours and 1 independent group of untreated cells. Normalized gene expression (-CT) within SMAD4 wild-type and SMAD4 knockout group of cells per each gene was used to generate the clustergram with heatmap. The colour scale indicates -CT values. (A, B) The arithmetic mean of housekeeping genes ACTB, B2M, GAPDH, HPRT1 and RPLPO was used to calculate CT values for each gene.