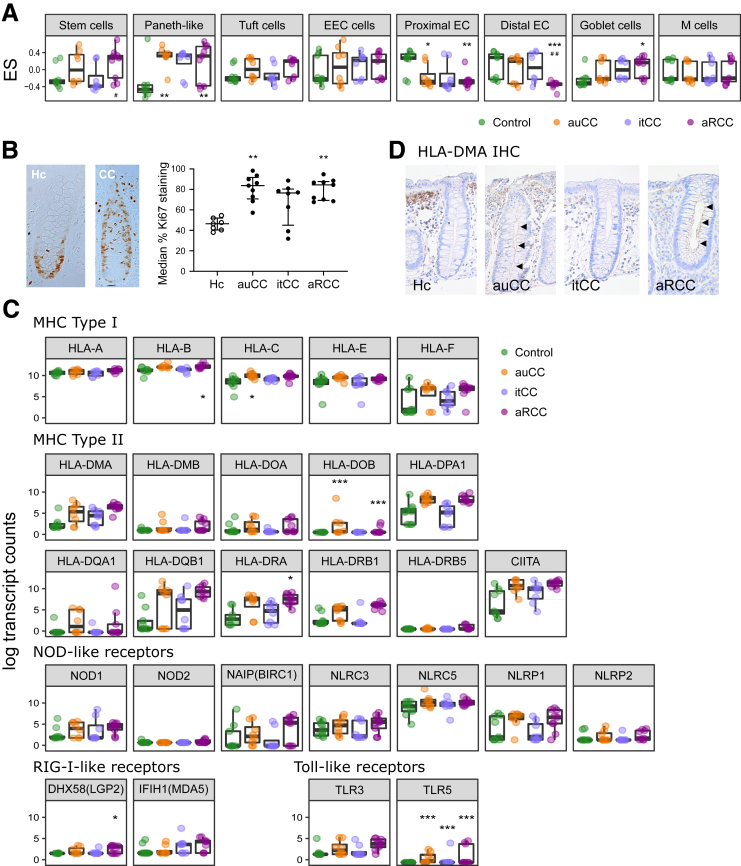
Figure 11.
CC IECs’ transcriptional profile suggest cell proliferation and recognition of antigens better than IEC from control subjects. (A) GSVA computed for different epithelial cell types from RNA-seq data displaying enrichment scores (ES) (median with interquartile range). Hc subjects are shown in green, auCC samples in orange, itCC samples in blue, and aRCC samples in purple. (B) Representative IHC images of longitudinally sectioned epithelial glands stained for Ki67 proliferation marker (brown) in paraffin-embedded sections of Hc and CC colonic mucosa (left). Analysis of Ki67 relative staining to total crypt length is shown on the right (median with interquartile range, median of 9 crypts/patient). (C) Pattern recognition receptor and HLA gene expression in intestinal epithelial cells from CC mucosa as normalized log2-transformed fold changes (using the regularized log function in R) of RNA-seq transcript counts (median with interquartile range) of pattern recognition receptors including members from the NOD-like, RIG-I-like, and Toll-like families, and HLA genes (divided into genes coding for major histocompatibility complex [MHC] type I and II proteins). Group colors are the same as in A. (D) Representative IHC images of HLA-DMA staining of paraffin-embedded sections from Hc, auCC, itCC, and aRCC colonic samples. Note the brown staining in the apical side of IEC sin crypts from active samples (black arrows). n = 7–13 samples per group. Statistically significant differences relative to Hc samples are shown as ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001; statistically significant differences relative to itCC samples are shown as #P < .05 and ##P < .01. EEC cells, enteroendocrine cells; EC, enterocytes; M cells, microfold cells.