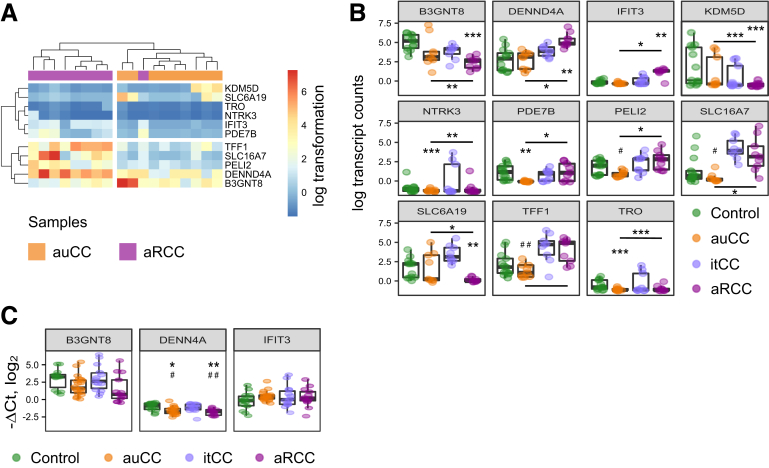
Figure 7.
DEGs between auCC and aRCC colonic mucosa. (A, B) Normalized log2-transformed fold changes (using the regularized log function in R) of RNA-seq transcript counts (log TC) from DEGs between auCC and aRCC CC mucosa (A) displayed as a heatmap or (B) as detailed individual plots including samples from Hc subjects and all CC groups (median with interquartile range). Selected genes for further RT-qPCR validation are highlighted in red. (C) Log2 fold changes (–ΔCt values) (median with interquartile range) in gene expression of B3GNT8, DENN4A, and IFIT3 analyzed by quantitative PCR. HPRT1 was used as a housekeeping control. Hc subjects are shown in green, auCC samples in orange, itCC samples in blue, and aRCC samples in purple. n = 9–13 samples per group for RNA-seq analyses; n = 13–20 samples per group for RT-qPCR validation. Statistically significant differences relative to Hc samples are shown as ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001, unless other comparison is indicated; statistically significant differences relative to itCC samples are shown as #P < .05, and ##P < .01.