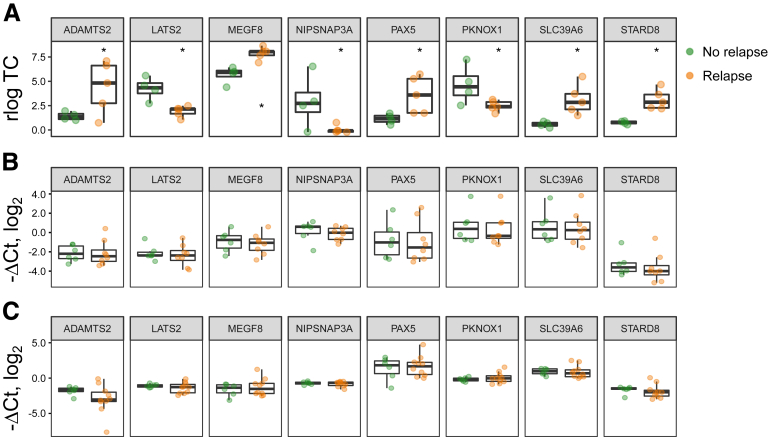
Figure 9.
DEGs in itCC samples from patients that experienced a disease relapse against those who did not. (A) Normalized log2-transformed fold changes (using the regularized log function in R) of RNA-seq transcript counts (log TC) (median with interquartile range) of DEGs between itCC samples from patients that experienced a disease relapse (orange), or did not (green). n = 4–5 samples per group. (B, C) Log2 fold changes (–ΔCt values, median with interquartile range) in gene expression from DEGs identified by RNA-seq in (B) colonic samples or (C) blood of itCC patients that suffered from a disease relapse (orange), or did not (green), as analyzed by RT-qPCR. HPRT1 was used as a housekeeping control. n = 6–10 samples per group for validation analyses. All primers detect all coding transcript variants of the indicated gene. n = 4–5 samples per group. Statistically significant differences are shown as ∗P < .05.