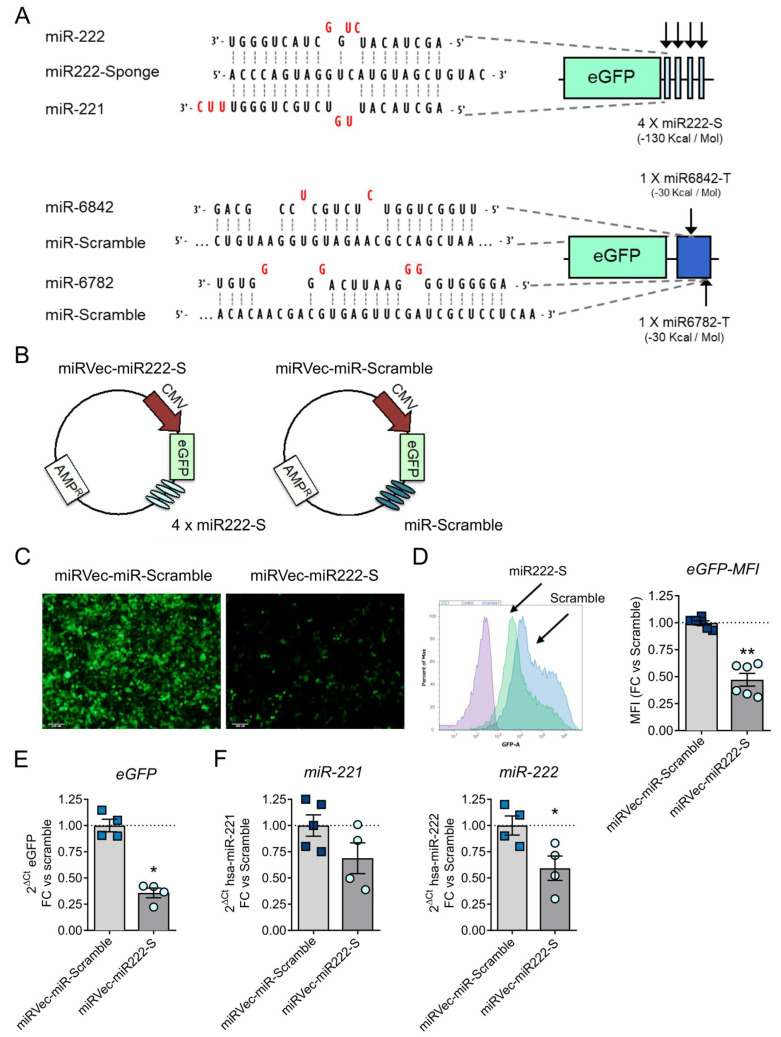
Figure 3.
Generation and functional validation of miR-222 sponge in miRVec-eGFP-miR222-S and miRVec-eGFP-Scramble. (A). Scheme of the sponge design with a sequence alignment of miR-222, miR-221, and the sponge (upper figure). A representation of the Scramble sequence with predictive off-targets miRNAs is also provided (lower figure). (B). Schematic representation of miRVec-eGFP-miR222-S and miRVec-eGFP-Scramble constructs. (C–F). Expression of eGFP from HEK-293T cells transfected with miRVec-eGFP-miR222-S or miRVec-eGFP-Scramble. (C). Representative images of eGFP fluorescence (scalebar 100 µm). (D). eGFP quantification by flow cytometry. The left panel shows a representative plot. The right panel shows the quantification of mean fluorescence intensity (MFI). (E). eGFP quantification by qRT-PCR. (F). Quantification of miR-221 and miR-222 content in HEK-293T cells transfected with miRVec-eGFP-miR222-S and miRVec-eGFP-Scramble. Values are represented as fold change compared to the Scramble construct. Data are shown as mean ± SEM for at least four independent biological replicates. Significance was assessed using a two-tailed Mann–Whitney test. * p < 0.05, ** p < 0.01.