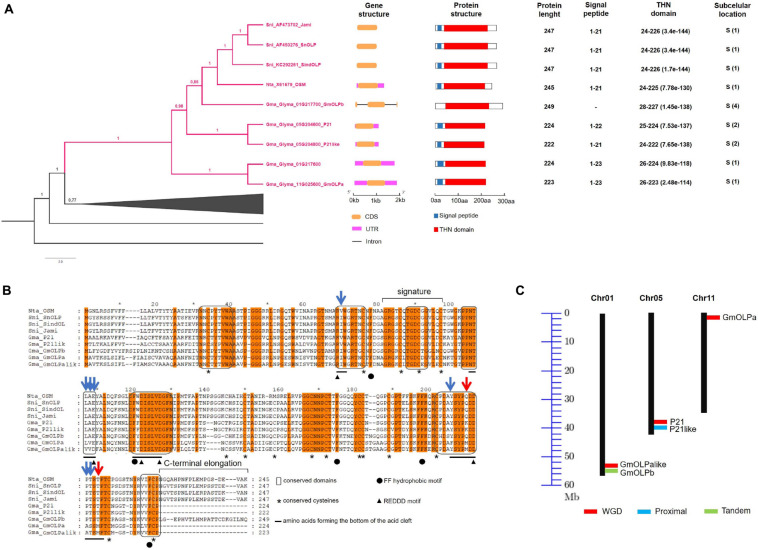
FIGURE 1.
Osmotin in silico analysis. (A) Detailed representation of the phylogenetic relationship among the thaumatin domain within the osmotin monophyletic clade with corresponding gene and protein structure information. According to the phylogenetic results the Gma_Glyma_01G217600 protein was named GmOLPa-like. Subcellular locations are numbered according to the TargetP server from one to five, where one indicates the strongest prediction. S, secretory pathway. (B) Alignment of osmotin protein sequences. Conserved residues and protein secondary structure are indicated according to Petre et al. (2011). Orange letter background corresponds to conserved amino acids (aa) among the nine analysed sequences. Blue and red arrows indicate aa substitutions with similar and different properties, respectively. (C) Chromosomal location and duplication of soybean osmotins. Black vertical lines represent the chromosomes with their numbers at the top. WGD, whole-genome duplication.