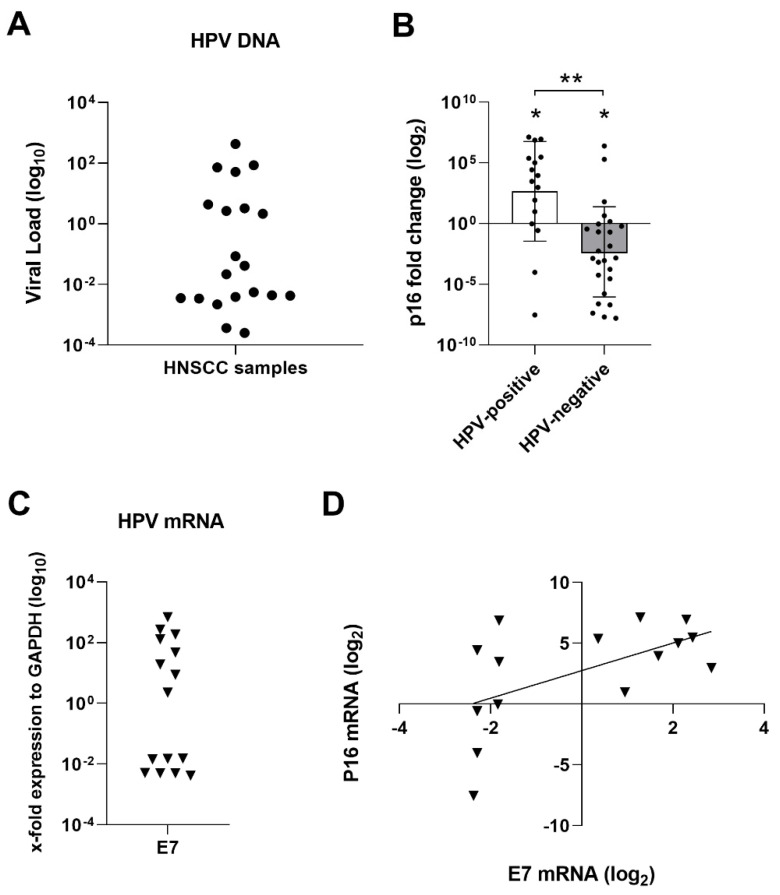
Figure 1.
Analysis of classical markers for stratification of HNSCC samples. Statistical significance was indicated as * for p < 0.05 and ** for p < 0.0001; (A) viral load quantification of HPV-positive HNSCC samples by qPCR; (B) differential p16 mRNA expression in HNSCC samples analyzed by qPCR; (C) Viral E7 mRNA expression in HPV-positive HNSCC samples; (D) Spearman correlation analyses between the expression of E7 (log2) oncogene and p16 (log2) showed correlation (r = 0.59; p < 0.05) in HPV-positive HNSCC tumor samples.