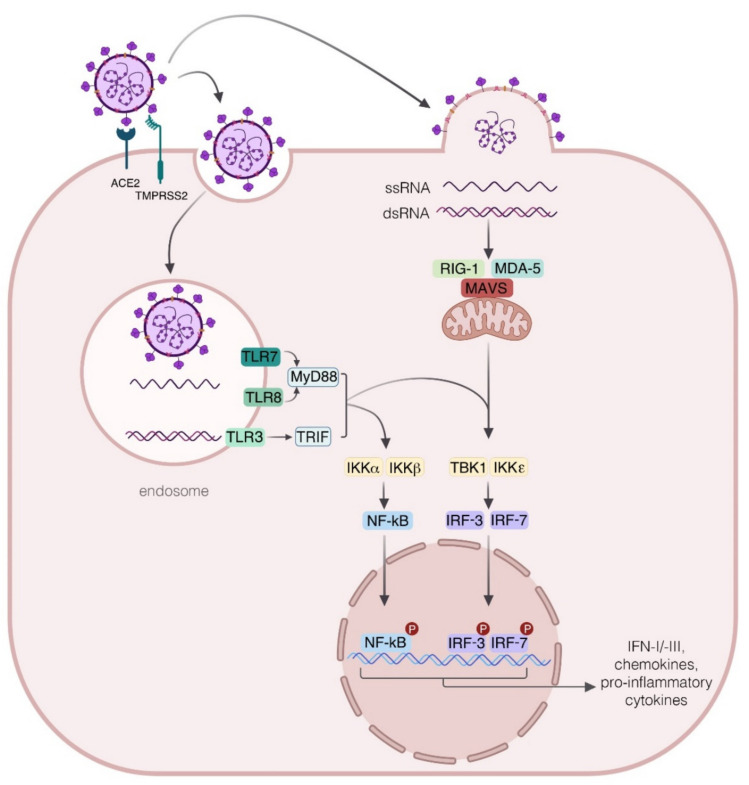
Figure 1.
Innate immune response to SARS-CoV-2. The interaction of SARS-CoV-2 with targeted cells requires the recognition of viral spike (S) protein by the angiotensin-converting enzyme (ACE2) and the priming of the S protein by the host cell transmembrane serine protease 2 (TMPRSS2). SARS-CoV-2 can gain access to host cells by means of the endocytic pathway that allows virus localization into the endosomes, where the viral single-stranded (ss) RNA is detected by the endosomal Toll-like receptors (TLR)7 and 8. Soon after viral RNA recognition, the adapter protein myeloid differentiation factor 88 (MyD88), is engaged for the activation and nuclear translocation of transcriptional factors nuclear factor kappa light-chain-enhancer of activated B cells (NF-κB) and interferon regulatory factor-3 (IRF-3) and 7 (IRF-7) for the transcription of genes encoding for proinflammatory cytokines and type I and III interferons (IFNs). In addition, TLR3 can be activated by double stranded (ds) RNA, produced during viral replication, thus contributing via the TRIF pathway to a protective response in SARS-CoV-2 infection. The sensing of replicating virus also occurs by the cytosolic receptor retinoic acid-inducible gene I (RIG-I) and the melanoma differentiation-associated protein (MDA5), which in turn recruits the mitochondrial antiviral signaling protein (MAVS), then leading to the IRF-3 and IRF-7 phosphorylation required for the expression of IFNs. IKKα: I-Kappa-B Kinase Alpha; IKKβ: I-Kappa-B Kinase Beta; IKKε: I-Kappa-B kinase epsilon; TBK1: TANK binding kinase 1; TRIF: TIR domain-containing adapter protein inducing interferon (IFN)-β.