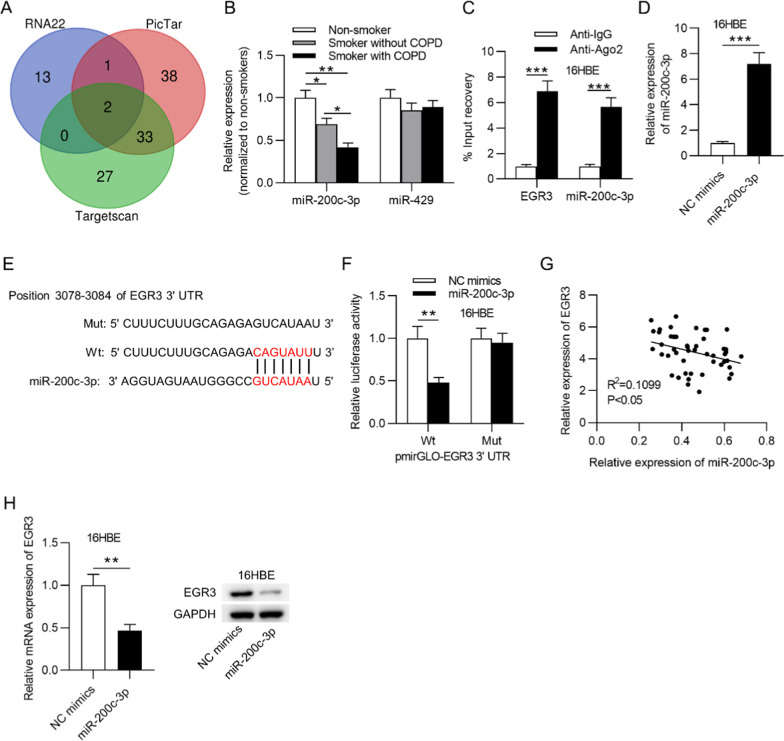
Fig. 3.
EGR3 was a downstream target of miR-200c-3p. a Venn diagram showing the potential miRNAs possessing binding sites on the EGR3 3’UTR. b RT-qPCR was used to examine the expression of miR-200c-3p and miR-429 in lungs from nonsmokers (n = 7), smokers without COPD (n = 22), and smokers with COPD (n = 26). c RIP assay was conducted to reveal the relative enrichment of miR-200c-3p and EGR3 precipitated by Ago2 normalized to that by IgG in 16HBE cells (n = 3). d RT-qPCR was used to measure miR-200c-3p expression in 16HBE cells transfected with NC mimics or miR-200c-3p mimics (n = 3). e The starBase database predicted the potential binding site of miR-200c-3p on the EGR3 3’UTR. f Luciferase reporter assay was used to assess the binding ability of miR-200c-3p and EGR3 in 16HBE cells (n = 3). g The correlation between EGR3 and miR-200c-3p expression in lung tissues of smokers (n = 48) was analyzed by Pearson’s correlation analysis. h The mRNA and protein levels of miR-200c-3p in response to miR-200c-3p overexpression in 16HBE cells (n = 3). *p < 0.05, **p < 0.01, ***p < 0.001