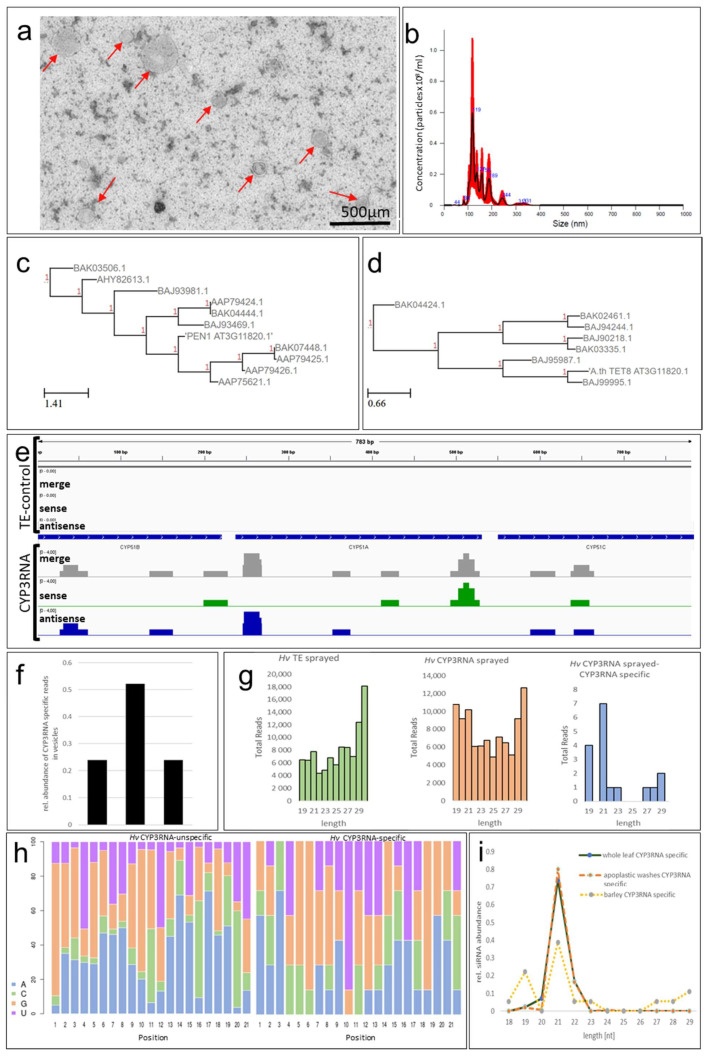
Figure 1.
(a) Barley EVs were negatively stained onto copper formvar meshes using 2% uranyl acetate. (b) Next, 5 µL of purified EVs was diluted up to a volume of 500 µL. The vesicle suspension was loaded onto a Nanosight NS300 (Malvern Panalytical). Five measurements were performed at 25 °C, and size, concentration prediction, and statistical analyses were performed using the NTA 3.2 Dev Build 3.2.16 software. (c,d) Arabidopsis thaliana PEN1 (AT3G11820) and TET8 (AT2G23810) paralogs of Hordeum vulgare subsp. vulgare (Hv) were predicted by the NCBI’s protein BLAST service (Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 14 June 2021)) and visualized using the ete view tool. Available online: http://etetoolkit.org (accessed on 23 June 2021). (e) RNA was isolated from mock and dsRNA-treated barley leaves. Indexed sRNA libraries were pooled and sequenced on the Illumina MiSeq Platform (1 × 36 SE). The readings were then mapped onto the CYP3RNA sequence. (f) The relative abundance of reads aligned to each CYP3RNA fragment (CYP51A, CYP51B, CYP51C) were calculated and (g) reads were sorted based on their size. (h) The nucleotide distribution for every position was counted for the 21 nts long siRNAs of all barley siRNAs (left) and siRNAs with perfect complementarity towards the CYP3RNA precursor (right). (i) The relative abundances of siRNA with different lengths from barley EVs were compared with relative siRNA abundances from Arabidopsis EVs purified from apoplastic washes and Arabidopsis vesicles isolated by whole-leaf vesicle purification.