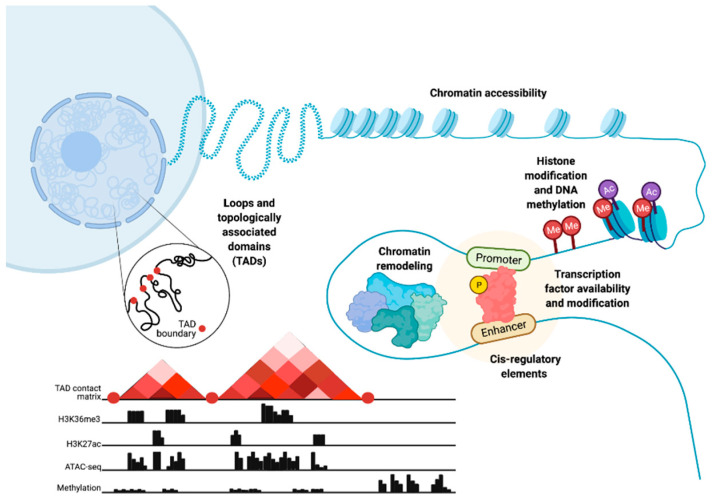
Figure 1.
Epigenetic- and chromatin-related mechanisms with potential for dysregulation in prostate cancer cells. Epigenetic dysregulation can occur at multiple levels, including changes in chromatin accessibility, histone and DNA modification through processes such as methylation, chromatin remodeling, modification of transcription factors and changes in their availability, cis-regulatory elements, chromatin loops, and topologically associated domains. These chromatin and epigenetic features can be analyzed via the integration of multiple high-throughput sequencing data types. These data include chromatin conformation capture (Hi-C) to understand the 3D chromatin structure and topologically associated domains, chromatin immunoprecipitation (ChIP-seq) to study histone markers, assay for transposase-accessible chromatin (ATAC-seq) to show chromatin accessibility patterns, and DNA methylation sequencing. Figure created with BioRender.com.