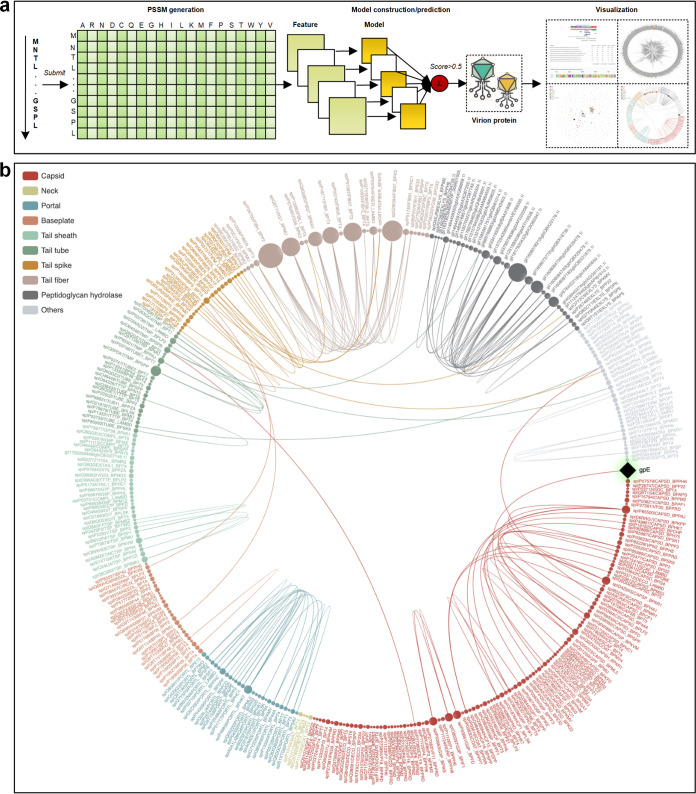
FIG 1.
Construction and workflow for STEP3. (a) Graphic summarizing the construction and prediction process of STEP3. A set of experimentally validated virion proteins and nonvirion proteins was compiled, and sequence data were fed into five PSSM models, including AAC-PSSM (59), PSSM composition (60), DPC-PSSM (59), AADP-PSSM (59), and a MEDP (61) model. The five individual models were trained based on five balanced subsets, and their prediction scores were averaged to obtain an ensemble model. Finally, five baseline models (corresponding to five evolutionary features) were further integrated as the final ensemble model of STEP3 through averaging their prediction scores. Support vector machine (SVM) with a radial basis function kernel was used to train each model. This ultimately provides a prediction of a “virion protein” which would be a structural component of the phage virion. (b) STEP3 data visualization provides a means to document relationships between a protein of interest. The example given is the protein component gpE from phage λ, which shows clear similarity to major capsid proteins from other phages. Structural studies confirm that despite limited sequence similarity, gpE is part of a family of major capsid proteins (9). Alternative visualization features are available in STEP3 (see Fig. S1 in the supplemental material).