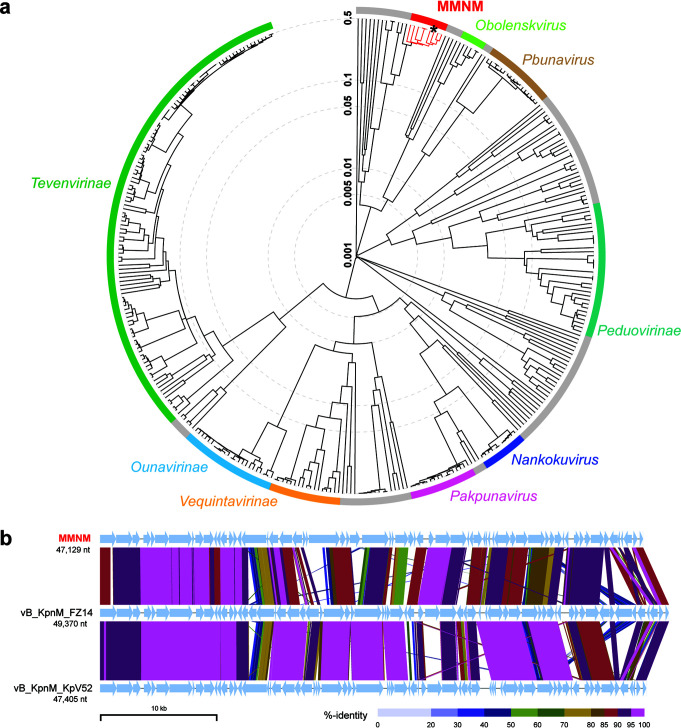
FIG 4.
Comparative genome analysis of Klebsiella phage MMNM. (a) Proteomic tree analysis of Myoviridae that infect Gammaproteobacteria. The branch lengths represent genomic similarity based on normalized pairwise sequence similarity scores plotted on a logarithmic scale. The tree was constructed using sequences from the default ViPTree data set and the following selected Klebsiella phage genomes: vB KpnM KpV79 (GenBank accession no. NC_042041), vB KpnM FZ14 (MK521906), vB KpnM KpV52 (NC_041900), 1611E-K2-1 (MG197810), vB KpnM IME346 (MK685667), vB KpnM 15-38 KLPPOU148 (MN689778), PEAT2 (NC_044940), and MMNM (MT894004). Viral subfamilies or genera are indicated by the colored bars. Gray bars represent phages that are currently unclassified. All known members of the Jedunavirus, including Klebsiella phage MMNM (*), are highlighted in red. (b) Whole-genome alignment of Klebsiella phage MMNM, vB_KpnM_FZ14, and vB_KpnM_KpV52. Each genome has been oriented to start with the gene encoding the putative tape measure protein. The sequences are linked by colored bars highlighting sequence identity values as shown in the key.