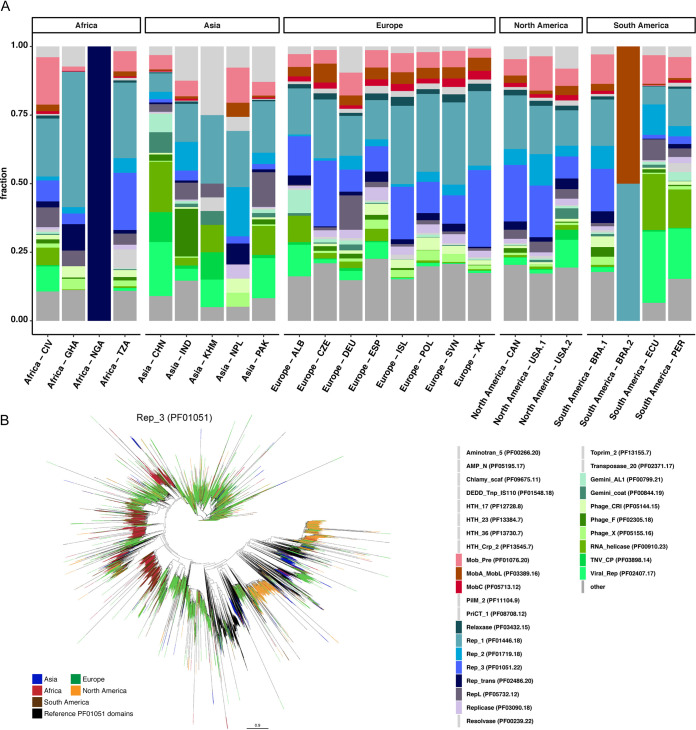
FIG 2.
Functional characterization of circular DNA elements based on protein families. (A) Bar plot displaying the fraction of Pfam identifiers assigned to predicted proteins on the circular elements. The 31 Pfam identifiers represent the Top10 Pfam identifiers for each sample. Protein domains specifically involved in plasmid mobilization and plasmid replication are indicated in red and blue, respectively (see legend to the bottom right). Virus/phage-related Pfam identifiers are indicated in green. Remaining Pfam identifiers are grouped (other) and are indicated in dark gray. (B) Data for proteins with a Rep_3 (PF01051) domain (n = 24,824) were combined with the 1,637 reference Rep_3 (PF01051) proteins from Pfam. The protein sequences with a length of ≥40 amino acids (n = 16,930) were aligned using MAFFT. A phylogenetic tree was build using FastTree and visualized using FigTree. A high-resolution version of the phylogenetic tree is available from Figshare at https://doi.org/10.6084/m9.figshare.14112992.