ABSTRACT
Pattern recognition receptors (PRRs) form the front line of defense against pathogens. Many of the molecular mechanisms that facilitate PRR signaling have been characterized in detail, which is critical for the development of accurate PRR pathway models at the molecular interaction level. These models could support the development of therapeutics for numerous diseases, including sepsis and COVID-19. This review describes the molecular mechanisms of the principal signaling interactions of the Toll-like receptor, STING, MAVS, and inflammasome pathways. A detailed molecular mechanism network is included as Data Set S1 in the supplemental material.
KEYWORDS: pattern recognition receptor, pathogen-associated molecular pattern, damage-associated molecular pattern, Toll-like receptor, inflammasome
INTRODUCTION
Pattern recognition receptor (PRR) pathways are highly diverse and include the Toll-like receptors (TLRs), C-type lectin receptors, NOD-like receptors, RIG-I-like receptors, and AIM2-like receptors (1, 2). PRR pathway mechanisms range from simple bimolecular association reactions to intricate reorganizations of supramolecular organizing centers (SMOCs). Clinically, PRR pathways can be activated by a single ligand (e.g., by an adjuvant), by a single pathogen (e.g., SARS-CoV-2), or by a highly diverse population of microbes (e.g., septic shock caused by acute appendicitis and opportunistic infections in immunocompromised individuals). PRR pathways can also be chronically activated in the absence of microbes (e.g., inflammaging) (3–5). Accurate modeling of PRR pathways is necessary to support the development of prophylactics, diagnostics, and therapeutics to fight numerous diseases related to the innate immune system.
PRR ligands include pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs). Human PRRs have been reported to form 89 distinct ligand-PRR interactions (PRRDB v2; downloaded 2 June 2021; interaction in ≥2 entries; only ligands of natural origin; https://webs.iiitd.edu.in/raghava/prrdb2/) (6). Of these, 67 involved TLRs. Signaling downstream of PRRs is notable for its utilization of numerous SMOCs (described below). PRR pathways often terminate at the level of transcriptional reprogramming, cytokine release, and programmed cell death (2).
Numerous investigators have published detailed molecular interaction networks of the mammalian TLR pathways (7–15). In addition, many online pathway databases (e.g., the Kyoto Encyclopedia of Genes and Genomes) describe molecular interactions within pathways of the innate immune system. This review provides significantly more detail by describing the molecular reactions corresponding to each interaction. For example, a simple protein-protein interaction might involve only two molecular reactions: one association and one dissociation. In contrast, formation of the myddosome involves numerous copies of the core proteins (six MyD88, four IRAK4, and four IRAK1/2) and multiple instances of phosphorylation between the complexed IRAKs (described below).
We present a detailed review of the TLR pathway (Fig. 1), as well as the STING, MAVS, inflammasome, and interferon pathways, in a systematic way that will facilitate modeling. In part, we selected these pathways because they are among those that have known roles in the pathogenesis of bacterial and viral sepsis (2, 16–19). In addition, these pathways have been well characterized mechanistically (described below).
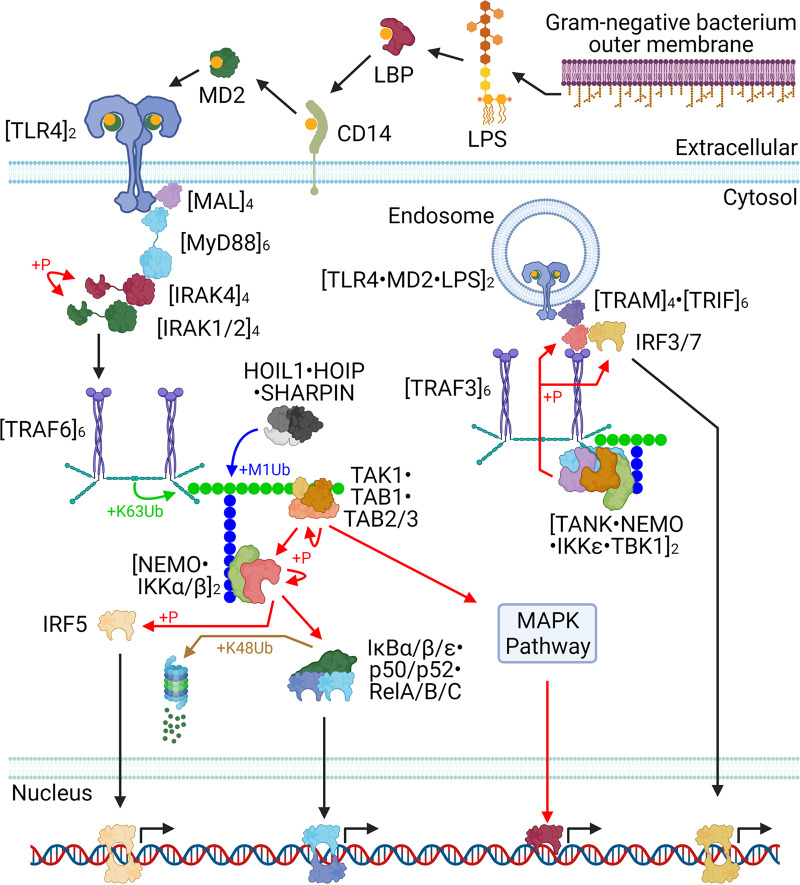
FIG 1.
The core TLR4 pathway. The key interactions of both the extracellular (MyD88-dependent) and endosomal (MyD88-independent) components of the TLR4 pathway are illustrated. Note that it is unclear if TRAF3 K63-polyubiquitin chains undergo M1 polyubiquitin branching, but if so, NEMO probably binds to them.
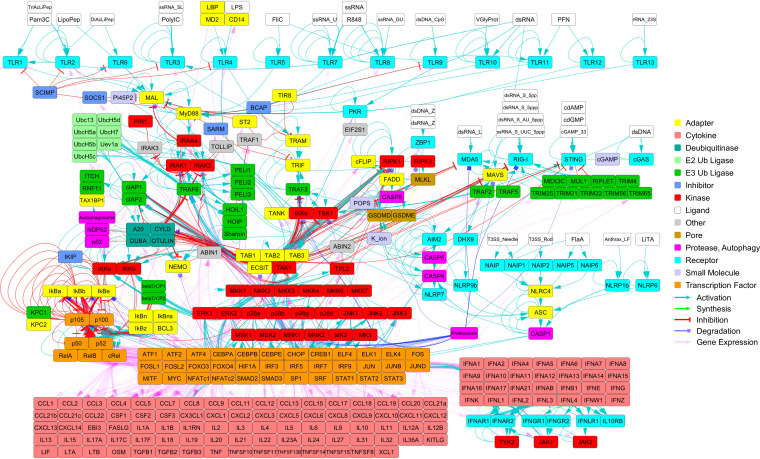
A tabulated molecular mechanism network is included as Data Set S1 in the supplemental material. Each molecular mechanism was manually curated (peer-reviewed articles were cited), and each mechanism was described in detail (e.g., protein complex requirements and phosphorylation states). The network includes both the human and mouse molecular mechanisms. Data Set S1 can be imported into a wide variety of network tools such as Cytoscape (https://cytoscape.org/) (20). With additional data (e.g., molecular reaction rates such as kon, koff, and kcat), all or part of Data Set S1 could be used to construct an SBML model (21) and/or used to perform pathway simulations using, for example, Simmune (22). Illustrations of the whole network (Fig. 2), a TLR4 network (Fig. S1), and a cytosolic PRR network (Fig. S2) were prepared. In addition, a summary of the network (Table 1) and a breakdown of the network by module (Table 2) are below.
FIG 2.
The PRR pathway network. The PRR network (see Data Set S1) is depicted. The network includes both the human and the mouse interactions. Two subnetworks of this network (the TLR4 pathway and the cytosolic PRR pathways) are shown in Fig. S1 and S2.
TABLE 1.
Summary of the molecular mechanism network
| Attribute | Count |
|---|---|
| Modules | 18 |
| Molecules | 398 |
| Proteoforms | 312 |
| Ligands (and related) | 39 |
| Systems (e.g., proteosome) | 47 |
| Interactions | 2,687 |
| Reactions | 3,855 |
| References | 253 |
| Citations | 5,785 |
TABLE 2.
Summary of the interactions within each module
| Module | Interactions |
|---|---|
| PAMP cell entry | 51 |
| TLR | 70 |
| Myddosome | 56 |
| K63-polyUb | 109 |
| IKK | 79 |
| NF-κB | 269 |
| MAPK | 159 |
| IRF | 34 |
| RIPK | 49 |
| STING | 18 |
| PKR | 8 |
| MAVS | 36 |
| Autophagy | 52 |
| Deubiquitinase | 103 |
| Inflammasome | 53 |
| Gene expression | 1,340 |
| Secretion | 109 |
| IFN | 92 |
Molecular reaction network of core elements of the TLR4 pathway. Instances of multiple edges between nodes indicates that there are multiple molecular interaction mechanisms. For example, the NF-κB proteins (p105, p100, p50, p52, RelA, RelB, and cRel) can dimerize, and each dimer can associate with an IκB protein (IκBa, IκBb, IκBe, p105, p100; the other IκB proteins are not shown). Download FIG S1, PDF file, 1.1 MB (1.1MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Molecular reaction network of core elements of the cytosolic PRR pathways. The MAVS and STING pathways are on the left, and the inflammasomes are on the right. The pathway diagram spans only from the PAMPs to selected major effectors (the TRAFs, the transcription factors, and GSDMD). MAVS is active at the cytosolic side of the outer mitochondrial membrane, and STING is active at the cytosolic side of the ER membrane. Download FIG S2, PDF file, 0.6 MB (660.9KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
(Tab A) Ligands, small molecules, and “System” molecules. (Tab B) Proteoforms. (Tab C) Bimolecular interactions. Download Data Set S1, XLSX file, 0.2 MB (248.5KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The network includes transcription factor-target gene relationships for cytokines and PRR pathway proteins. These could support the modeling of transcription and translation and enable longer (in silico time) pathway simulations. These transcription factor-target gene relationships were constructed using data from two online databases. CytReg (downloaded 2 June 2021; http://cytreg.bu.edu) (23) was used to model transcription factor-mediated expression of cytokines. RegNetwork (downloaded 2 June 2021; http://www.regnetworkweb.org) (24) was used to model transcription factor-mediated expression of PRR pathway proteins (except for cytokines). Proteins were not included if they were outside the PRR pathways (e.g., metabolic enzyme expression affected by the mitogen-activated protein kinase [MAPK]-activated transcription factors).
TLR PATHWAY
TLRs are a class of dimeric PRRs which sense a variety of extracellular and endosomal PAMPs and DAMPs and initiate signal transduction resulting in transcriptional reprogramming or programmed cell death (25–28). The vertebrate TLR family of genes contains at least 27 members, of which 13 are found in the mammalian TLR family (29). Humans express TLR1 to TLR10, whereas mice express TLR1 to TLR9 and TLR11 to TLR13 (29). One or more PAMPs have been discovered for all 13 mammalian TLRs, and DAMPs have been discovered for almost all 13 (29–34).
Human TLRs have been reported to bind 46 distinct ligands of natural origin (PRRDB v2; downloaded 2 June 2021; interaction in ≥2 entries; https://webs.iiitd.edu.in/raghava/prrdb2/) (6). Many TLR-ligand structures have been solved (35–38). Some TLR dimers bind to only a single ligand copy. For example, the TLR1-TLR2 heterodimer binds to only a single copy of triacyl-lipopeptide (35).
TLR activation initiates a signal transduction which includes the MAPK phosphorylation cascade and the NF-κB pathway (25–28). This results in broad reprogramming of gene expression, resulting in physiological alterations related to metabolism, cell survival, cell stress, and cytokine signaling (25–28). Numerous pathogens express virulence factors that antagonize signaling proteins of the TLR pathway (39).
TLR4 activation.
Lipopolysaccharide (LPS) is a highly diverse class of molecules consisting of a lipid moiety, a core oligosaccharide moiety, and an O-antigen moiety (40–42). The chemical structure of an LPS molecule determines its function. For example, Escherichia coli LPS is proinflammatory whereas Yersinia pestis LPS is anti-inflammatory after temperature-specific remodeling at 37°C (41). Though energetically unfavorable, LPS sheds from bacterial outer membranes, binds LPS-binding protein (LBP), and then associates with CD14 (LBP of the LPS-LBP binds CD14, and then this complex reorganizes until LPS-CD14 releases LBP) (42–44). CD14-bound LPS translocates to MD2, and LPS-MD2 binds a TLR4 monomer (43, 44). Two copies of LPS-MD2-TLR4 can associate at their ectodomains and subsequently associate at their Toll/interleukin-1 (IL-1) receptor (TIR) domains (43–45). A ligand-bound TLR4 cannot associate with an unbound copy at their ectodomains (45).
Myddosome formation.
The TIR domain of every mammalian TLR dimer except TLR3 can activate MyD88 and stimulate the formation of a myddosome (30). TLR1, TLR2, TLR4, TLR6, and TLR9 first bind to TIR domain-containing adapter protein (MAL), and then the MAL TIR domain binds to and activates MyD88 (30, 46, 47). The MAL TIR domain can oligomerize, and it is unclear if these oligomers can cause TLR clustering (46–48). The MyD88 TIR domain can oligomerize beyond six subunits in vitro, but in a functional myddosome it may be limited to six copies (25, 46, 47, 49, 50). Each MyD88 TIR domain is tethered to its death domain (DD), and the MyD88 hexamer DDs bind four copies of the interleukin-1 receptor-associated kinase 4 (IRAK4) via its DDs (25, 46, 47, 49). Likewise, these four IRAK4 DDs each bind to either an IRAK1 DD or an IRAK2 DD (25, 46, 47, 49). The core myddosome is therefore composed of 6:4:4 copies of MyD88:IRAK4:[IRAK1 or IRAK2].
The IRAK4 kinase domains (KDs) catalyze intracomplex phosphorylation of each other (51). The phospho-IRAK4 KD binds to and phosphorylates an IRAK1 DD or an IRAK2 DD (49, 51). Eventually, the IRAK1/2 copies each become multiply phosphorylated, which destabilizes the myddosome (25, 49, 52). The IRAK1/2 copies are released (possibly still bound to IRAK4 and Myd88) (25, 52).
K63-M1 polyUb signaling.
Active IRAK1/2 tetramers bind and activate the TRAF-C domain of tumor necrosis factor (TNF) receptor-associated factor 6 (TRAF6) monomers (53, 54). TRAF6 subsequently forms a homotrimer at the TRAF-C and coiled-coil (CC) domains (53, 54). TRAF6 trimers subsequently dimerize at their RING domain, forming an active homohexamer which can homo-oligomerize (53, 54). Activated TRAF6 is an E3 ubiquitin ligase which catalyzes K63-linked polyubiquitin (polyUb) onto numerous substrates including IRAK1, IRAK2, TRAF6, and NEMO (53–55). Subsequently, the LUBAC complex (composed of HOIP, HOIL1, and Sharpin) catalyzes M1-linked polyubiquitination branches off the K63-linked chains (55, 56).
TAB2 and TAB3 are components of a heterotrimer consisting of TAB1, TAK1, and either TAB2 or TAB3 (55). TAK1 kinase activity is activated by TAB2/3 binding to K63-polyUb (55, 57). TAK1 autophosphorylates itself (54) and initiates MAPK signaling by phosphorylating and activating MKK3, MKK4, MKK6, and MKK7 (58). These MKKs phosphorylate and activate p38α, p38β, p38γ, p38δ, JNK1, JNK2, and JNK3 (58). These MAPKs activate numerous transcription factors, broadly reprogramming transcription and affecting metabolism, growth, proliferation, programmed cell death, stress, immunity, chemotaxis, and differentiation (58).
In addition, TAK1 phosphorylates nearby substrates that are also bound to K63-M1 polyubiquitin. NEMO (at its ubiquitin binding domain) binds to M1-polyUb (53–55, 57, 59), and this step is critical for TAK1 activation of IκB kinase alpha (IKKα) and IKKβ (IκB is short for inhibitor of NF-κB) and subsequent NF-κB pathway activation. IKKα, IKKβ, and NEMO each form homodimers, and a NEMO homodimer (via its kinase binding domain) can bind to either an IKKα homodimer or an IKKβ homodimer (via their NEMO binding domains) (53, 54, 59). Additionally, the IKKα and IKKβ homodimers can form higher-order homomeric complexes, and possibly also heteromeric complexes with each other. IKKα homodimers can trimerize (i.e., 6 copies total), and this complex can dimerize (i.e., 12 copies total; higher-order IKKα structures were not reported) (60). IKKβ homodimers can dimerize (i.e., 4 copies total), trimerize (i.e., 6 copies total), and form a variety of higher-order oligomers (61). It remains unclear exactly which IKKα-IKKβ-NEMO complexes form in vivo.
As mentioned above, NEMO binds to M1-polyUb. This brings IKKα/β into close proximity with TAK1. TAK1 phosphorylates and activates IKKα and IKKβ (55, 56). Activation of IKKα/β is completed via intracomplex trans-autophosphorylation (55, 56). In addition to activating the NF-κB pathway, IKKβ also activates MAPK signaling via TPL2 (see Data Set S1 in the supplemental material for details), resulting in broad transcriptional reprogramming (58).
NF-κB pathway.
NF-κB is a class of five dimeric transcription factors: NF-κB1, NF-κB2, RelA, RelB, and c-Rel (54, 59, 62, 63). Unlike the three Rel forms, NF-κB1 and NF-κB2 lack a carboxy-terminal transactivation domain, and therefore, they cannot directly initiate transcription. Instead, the full-length forms of NF-κB1 and NF-κB2 (termed p105 and p100, respectively) each contain a carboxy-terminal ankyrin repeat domain (ARD). p105 processing into p50 is constitutive, whereas p100 processing into p52 is induced by its phosphorylation by IKKα (63). There are 28 NF-κB dimeric forms (Data Set S1).
The IκB family consists of nine members: p105, p100, IκBα, IκBβ, IκBε, IκBη, IκBζ, IκBNS, and BCL3 (59, 63). The IκB family members are all characterized by their ARD. The NF-κB family (including both forms of NF-κB1 and NF-κB2) all contain an amino-terminal Rel homology region (RHR) (54, 59, 62, 63). The IκB ARD regions can bind to NF-κB dimers (at the NF-κB RHR sequences) (54, 63). This results in exactly 200 potential trimeric complexes (Data Set S1; not every combination is necessarily chemically possible). The cytosolic IκB forms (p105, p100, IκBα, IκBβ, and IκBε) inhibit NF-κB by preventing its localization to the nucleus, whereas the nuclear IκB forms (IκBη, IκBζ, IκBNS, and BCL3) can function by activating or inhibiting transcription (54, 62, 63). Phosphorylation of the classical IκB forms (IκBα, IκBβ, and IκBε) by either IKKα or IKKβ results in dissociation of the IκB from the NF-κB RHR (54, 62, 63). This enables nuclear import of the NF-κB and K48 polyubiquitination and degradation of the IκB.
MyD88-independent TLR signaling.
TLR3 and TLR4 can signal independently of MyD88, and both do so exclusively from the endosome (25, 27). Activated TLR3 binds to and activates TIR domain-containing adapter molecule 1 (TRIF) directly (27, 64). In contrast, activated endosomal TLR4 binds to and activates TIR domain-containing adapter molecule 2 (TRAM) (27, 32, 65, 66). Subsequently, TRAM probably homodimerizes and/or homo-oligomerizes and then binds to and activates TRIF (32, 65–67). TRIF probably homodimerizes and/or homo-oligomerizes and binds to and activates TRAF3 and TRAF6 (27, 32, 65–67). Like TRAF6 (described above), activated TRAF3 forms a trimer, and then the trimers dimerize (i.e., six copies total), which enables its K63 E3 ubiquitin ligase activity (54). It is unclear if TRAF3 and/or TRAF6 can form heteromers with each other or with other TRAFs (68).
In addition to its K63-polyubiquitination signaling function, TRIF-bound TRAF3 also has a key role in signaling via IKKε and TBK1 (27). TRAF3 binds to dimeric TRAF family member-associated NF-κB activator (TANK), and then TANK binds to dimeric IKKε, dimeric NEMO, and dimeric TBK1 (66). TRAF3 probably K63-polyubiquitinates IKKε and TBK1, and then IKKε or TBK1 phosphorylates TRIF at its pLxIS motif (32, 69–72). The transcription factors IRF3 and (likely) IRF7 then bind to this motif and are phosphorylated and activated by IKKε or TBK1 (69, 73, 74). IRF5 is activated via a different mechanism; it is phosphorylated and activated by IKKβ (75). The three IRFs dimerize, translocate to the nucleus, and activate transcription of immune response genes (69, 74).
As described above, a key outcome of TLR activation is the reprogramming of gene expression (another TLR-mediated effect is programmed cell death, described below). Notably, TLR activation upregulates the production and secretion of cytokines (26, 64). For example, NF-κB upregulates the expression of interleukin-1 (IL-1), IL-2, IL-6, IL-8, IL-12, and TNF, which upregulate proliferation, inflammation, and angiogenesis (76). TLR activation also upregulates the production and secretion of interferons, which can act in an autocrine or paracrine manner, resulting in the transcriptional reprogramming of a wide range of genes with diverse effects (74, 77).
Programmed cell death.
Successful human pathogens often express virulence factors to antagonize the host immune response (78). For example, Yersinia pestis can enter host cells and secrete YopJ into the host cell cytosol which inhibits TAK1, IKKs, and MKKs, thereby blocking the NF-κB and MAPK pathways (78, 79). To prevent the host cell from becoming a means to immune evasion and/or pathogen proliferation, the TLR pathway activates receptor-interacting serine/threonine-protein kinase 1 (RIPK1), which will cause programmed cell death if it is not deactivated by the NF-κB and MAPK pathways.
Activated TRIF binds to RIPK1 as well as to TRAF6 (78). TRAF6 K63-polyubiquitinates and activates cIAP1 and cIAP2, which dimerize (70, 80, 81). cIAP1/2 K63-polyubiquitinates RIPK1 (78). Alternatively, activated TRIF binds to RIPK1, and RIPK1 homodimerizes and trans-autophosphorylates itself (82, 83). RIPK1 might first oligomerize and then trans-autophosphorylate itself (84–86). In a third option, RIPK1 can be phosphorylated and inhibited by numerous kinases (IKKα, IKKβ, IKKε, TAK1, TBK1, and MK2) under various circumstances (see Data Set S1 for details). Absent this inhibition, each monomer of the autophosphorylated RIPK1 homodimer binds to and activates a copy of fatty acid synthase (FAS)-associated death domain protein (FADD) (83, 87). Each of the two FADD copies binds to and activates a copy of caspase-8 (CASP8), and CASP8 trans-autoproteolyzes and activates itself (78, 83, 88–90). Activated CASP8 proteolyzes and activates gasdermin-D (GSDMD) and GSDME, which form pores, ultimately triggering apoptosis (89). Activated CASP8 also proteolyzes and activates the proinflammatory cytokines IL-1β and IL-18, which exit the cell through the pores (89). RIPK1 can alternatively trigger necroptosis via RIPK3 and MLKL (see Data Set S1 for details).
CYTOSOLIC PRR PATHWAYS
STING pathway.
Cyclic GMP-AMP synthase (cGAS) is an enzyme that is activated by binding to double-stranded DNA (dsDNA) originating from DNA viruses, retroviruses (via reverse transcription), bacteria, damaged mitochondria, phagocytosed dead cells, and host cell chromatin (e.g., via genomic instability and retrotransposons) (64, 91). Upon DNA binding, cGAS homodimerizes and synthesizes cyclic-GMP(2′-5′)-AMP(3′-5′), which binds to and activates STING (which is a homodimeric transmembrane protein located at the endoplasmic reticulum [ER] or Golgi complex) (64, 91). STING can also be activated by three bacterial cyclic dinucleotides: cyclic-di-AMP, cyclic-di-GMP, and 3′,3′-cyclic-GMP-AMP (92). Activated STING binds to homodimeric TBK1, and this complex oligomerizes (91). TBK1 trans-autophosphorylates and activates itself and then phosphorylates the STING pLxIS motif (69, 91). IRF3 binds to this motif and is activated via a similar mechanism as TRIF (described above) (69, 91).
MAVS pathway.
Long (>1,000-bp) double-stranded viral RNA binds to multiple copies of MDA5 (64, 93–95). Subsequently, TRIM25 and TRIM65 K63-polyubiquitinate MDA5, possibly causing it to homo-oligomerize (64, 94–96). MAVS is a transmembrane protein of the outer mitochondrial membrane, its CARD domain is cytosolic, and one or both of the CARD domains of MDA5 bind to the CARD domain of monomeric MAVS (64, 93–95). TRIM31 K63-polyubiquitinate MAVS, enabling it to oligomerize (96, 97). MAVS binds to and activates TRAF2, TRAF3, TRAF5, and TRAF6, which activate the NF-κB and MAPK pathways (described above and in Data Set S1 in the supplemental material) (64, 93, 96). In addition, IKKε and TBK1 phosphorylate the MAVS pLxIS motif, which ultimately results in the activation of IRF3 and IRF7 via a similar mechanism as TRIF and STING (described above) (64, 69, 93).
Short (10- to 300-bp) double-stranded viral RNA (5′ diphosphate or 5′ triphosphate) binds to single copies of RIG-I (64, 93, 95). RIPLET K63-polyubiquitinates the RIG-I C-terminal domain (CTD), and RIG-I homodimerizes (64, 95). MEX3C, RIPLET, TRIM4, and TRIM25 K63-polyubiquitinate the second RIG-I CARD domain (64, 93, 95, 96). RIG-I forms homotetramers and homomers, and the RIG-I CARD domains bind to and activate MAVS in a similar manner as MDA5 (64, 93, 95).
Inflammasomes.
Inflammasomes are cytosolic oligomeric sensors of numerous PAMPs. Inflammasomes promote proinflammatory cytokine signaling, they cause programmed cell death, and their structure and function have been recently reviewed in detail (46, 48, 98–108). Briefly, inflammasomes are composed of a receptor, an adapter, and an effector. Inflammasome ligand-receptor interactions include double-stranded DNA binding to AIM2, lipoteichoic acid binding to NLRP6, and lipopeptide binding to NLRP7. Some ligand-receptor interactions require an intermediate protein. For example, NLRP9b activation by double-stranded RNA requires DHX9, and NLRC4 activation by bacterial proteins (flagellin, and the rod and needle from type III secretion systems) requires NAIP. Finally, some ligands activate their receptor using a mechanism other than receptor ligation (e.g., Bacillus anthracis lethal factor cleaves NLRP1B).
Upon ligand binding, the receptor homo-oligomerizes (46, 48, 98–108). For example, NAIP-NLRC4 forms either a disk of ∼12 monomers (48, 98, 105, 106) or a helix of ∼12 monomers per turn (105). The receptor binds to the adapter protein ASC, which then homo-oligomerizes into a linear chain which might form branches (106). Two receptors (NLRC4 and NLRP1b) are capable of forming functional inflammasomes with or without ASC. Oligomeric ASC binds to CASP1, which homo-oligomerizes into a linear chain and is activated by trans-autoproteolysis. CASP1 processes and activates GSDMD, which forms pores in the cell membrane. CASP1 also processes the proinflammatory cytokines IL1β and IL-18, which exit the cell via the GSDMD pores. In contrast to the above canonical inflammasomes, the noncanonical inflammasome CASP4/5 directly binds to LPS, is activated by trans-autoproteolysis, and processes GSDMD. For more details, see Data Set S1.
DISCUSSION
The molecular mechanisms utilized by the TLR and cytosolic PRR pathways are highly diverse. Very characteristic are the oligomeric SMOCs and the branching chains of ubiquitin. It remains unclear what benefit, if any, is derived from the utilization of such large and complex molecular structures in the struggle against pathogens and their virulence factors. Modeling can be used to address these questions, but accurate modeling of these pathways remains challenging.
Molecular interaction networks of the TLR pathway have been published previously (7–14), and this review includes a molecular mechanism network of the TLR and cytosolic PRR pathways (see Data Set S1 in the supplemental material). Despite having significant scope and detail, none of these networks are comprehensive, primarily because so many significant pathway components remain obscure. For example, although numerous alternative splice isoform sequences of TLR pathway proteins have been sequenced, many of their functions remain unclear. This is unfortunate because it is likely that many of these isoforms have important physiological roles. For example, it is known that a truncated isoform of MyD88 is dominant negative and plays an important role in preventing chronic inflammation (109). Other unresolved issues include fully determining the structure-function relationship of branched polyubiquitin and identifying the phosphatases that act on TLR pathway phosphoproteins.
Developing PRR pathway models at molecular interaction level for performing pathway simulations will aid the development of therapeutics for diseases related to the innate immune system (e.g., sepsis), but developing these models will require more than just molecular mechanism networks. Critically, the molecular reaction rates (kon, koff, and kcat) and reactant concentrations are required. Targeted proteomics has been used to measure protein concentrations to support the development of numerous signaling pathway models (110). However, measuring the corresponding reaction rates remains challenging, but they can be predicted using bimolecular simulations (111, 112). These two sets of values can be used as input parameters for pathway modeling and simulation (22, 113). Importantly, pathway models can be trained using, for example, microscopy data, which is how we trained our model of the mouse macrophage chemotaxis signaling pathway (114). Although the development of PRR pathway models remains a formidable challenge, they will be necessary for accurately predicting the behavior of these pathways, especially for pathways stimulated by a diverse population of microbes and PAMPs.
ACKNOWLEDGMENT
This research was supported by the Intramural Research Program of the NIH, NIAID.
Contributor Information
Aleksandra Nita-Lazar, Email: nitalazarau@niaid.nih.gov.
Ileana M. Cristea, Princeton University
REFERENCES
- 1.Brubaker SW, Bonham KS, Zanoni I, Kagan JC. 2015. Innate immune pattern recognition: a cell biological perspective. Annu Rev Immunol 33:257–290. doi: 10.1146/annurev-immunol-032414-112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hotchkiss RS, Moldawer LL, Opal SM, Reinhart K, Turnbull IR, Vincent JL. 2016. Sepsis and septic shock. Nat Rev Dis Primers 2:16045. doi: 10.1038/nrdp.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fulop T, Larbi A, Dupuis G, Le Page A, Frost EH, Cohen AA, Witkowski JM, Franceschi C. 2017. Immunosenescence and inflamm-aging as two sides of the same coin: friends or foes? Front Immunol 8:1960. doi: 10.3389/fimmu.2017.01960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franceschi C, Garagnani P, Parini P, Giuliani C, Santoro A. 2018. Inflammaging: a new immune-metabolic viewpoint for age-related diseases. Nat Rev Endocrinol 14:576–590. doi: 10.1038/s41574-018-0059-4. [DOI] [PubMed] [Google Scholar]
- 5.Santoro A, Zhao J, Wu L, Carru C, Biagi E, Franceschi C. 2020. Microbiomes other than the gut: inflammaging and age-related diseases. Semin Immunopathol 42:589–605. doi: 10.1007/s00281-020-00814-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaur D, Patiyal S, Sharma N, Usmani SS, Raghava GPS. 2019. PRRDB 2.0: a comprehensive database of pattern-recognition receptors and their ligands. Database (Oxford) 2019:baz076. doi: 10.1093/database/baz076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.An GC, Faeder JR. 2009. Detailed qualitative dynamic knowledge representation using a BioNetGen model of TLR-4 signaling and preconditioning. Math Biosci 217:53–63. doi: 10.1016/j.mbs.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X, Zhong CQ, Yin Z, Qi H, Xu F, He Q, Shuai J. 2020. Data-driven modeling identifies TIRAP-independent MyD88 activation complex and myddosome assembly strategy in LPS/TLR4 signaling. Int J Mol Sci 21:3061. doi: 10.3390/ijms21093061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu B, Liu Q, Yang L, Palaniappan SK, Bahar I, Thiagarajan PS, Ding JL. 2016. Innate immune memory and homeostasis may be conferred through crosstalk between the TLR3 and TLR7 pathways. Sci Signal 9:ra70. doi: 10.1126/scisignal.aac9340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oda K, Kitano H. 2006. A comprehensive map of the toll-like receptor signaling network. Mol Syst Biol 2:2006.0015. doi: 10.1038/msb4100057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Padwal MK, Sarma U, Saha B. 2014. Comprehensive logic based analyses of Toll-like receptor 4 signal transduction pathway. PLoS One 9:e92481. doi: 10.1371/journal.pone.0092481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raza S, McDerment N, Lacaze PA, Robertson K, Watterson S, Chen Y, Chisholm M, Eleftheriadis G, Monk S, O’Sullivan M, Turnbull A, Roy D, Theocharidis A, Ghazal P, Freeman TC. 2010. Construction of a large scale integrated map of macrophage pathogen recognition and effector systems. BMC Syst Biol 4:63. doi: 10.1186/1752-0509-4-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wentker P, Eberhardt M, Dreyer FS, Bertrams W, Cantone M, Griss K, Schmeck B, Vera J. 2017. An interactive macrophage signal transduction map facilitates comparative analyses of high-throughput data. J Immunol 198:2191–2201. doi: 10.4049/jimmunol.1502513. [DOI] [PubMed] [Google Scholar]
- 14.Li F, Thiele I, Jamshidi N, Palsson BO. 2009. Identification of potential pathway mediation targets in Toll-like receptor signaling. PLoS Comput Biol 5:e1000292. doi: 10.1371/journal.pcbi.1000292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomaiuolo M, Kottke M, Matheny RW, Reifman J, Mitrophanov AY. 2016. Computational identification and analysis of signaling subnetworks with distinct functional roles in the regulation of TNF production. Mol Biosyst 12:826–838. doi: 10.1039/c5mb00456j. [DOI] [PubMed] [Google Scholar]
- 16.Grondman I, Pirvu A, Riza A, Ioana M, Netea MG. 2020. Biomarkers of inflammation and the etiology of sepsis. Biochem Soc Trans 48:1–14. doi: 10.1042/BST20190029. [DOI] [PubMed] [Google Scholar]
- 17.Wiersinga WJ, Leopold SJ, Cranendonk DR, van der Poll T. 2014. Host innate immune responses to sepsis. Virulence 5:36–44. doi: 10.4161/viru.25436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu Q, Knight PH, Ren Y, Ren H, Zheng J, Wu X, Ren J, Sawyer RG. 2019. The emerging role of stimulator of interferons genes signaling in sepsis: inflammation, autophagy, and cell death. Acta Physiol (Oxf) 225:e13194. doi: 10.1111/apha.13194. [DOI] [PubMed] [Google Scholar]
- 19.Lin GL, McGinley JP, Drysdale SB, Pollard AJ. 2018. Epidemiology and immune pathogenesis of viral sepsis. Front Immunol 9:2147. doi: 10.3389/fimmu.2018.02147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keating SM, Waltemath D, König M, Zhang F, Dräger A, Chaouiya C, Bergmann FT, Finney A, Gillespie CS, Helikar T, Hoops S, Malik-Sheriff RS, Moodie SL, Moraru II, Myers CJ, Naldi A, Olivier BG, Sahle S, Schaff JC, Smith LP, Swat MJ, Thieffry D, Watanabe L, Wilkinson DJ, Blinov ML, Begley K, Faeder JR, Gómez HF, Hamm TM, Inagaki Y, Liebermeister W, Lister AL, Lucio D, Mjolsness E, Proctor CJ, Raman K, Rodriguez N, Shaffer CA, Shapiro BE, Stelling J, Swainston N, Tanimura N, Wagner J, Meier-Schellersheim M, Sauro HM, Palsson B, Bolouri H, Kitano H, Funahashi A, Hermjakob H, Doyle JC, Hucka M, SBML Level 3 Community members . 2020. SBML level 3: an extensible format for the exchange and reuse of biological models. Mol Syst Biol 16:e9110. doi: 10.15252/msb.20199110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang F, Angermann BR, Meier-Schellersheim M. 2013. The Simmune Modeler visual interface for creating signaling networks based on bi-molecular interactions. Bioinformatics 29:1229–1230. doi: 10.1093/bioinformatics/btt134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carrasco Pro S, Dafonte Imedio A, Santoso CS, Gan KA, Sewell JA, Martinez M, Sereda R, Mehta S, Fuxman Bass JI. 2018. Global landscape of mouse and human cytokine transcriptional regulation. Nucleic Acids Res 46:9321–9337. doi: 10.1093/nar/gky787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu ZP, Wu C, Miao H, Wu H. 2015. RegNetwork: an integrated database of transcriptional and post-transcriptional regulatory networks in human and mouse. Database (Oxford ) 2015:bav095. doi: 10.1093/database/bav095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balka KR, De Nardo D. 2019. Understanding early TLR signaling through the Myddosome. J Leukoc Biol 105:339–351. doi: 10.1002/JLB.MR0318-096R. [DOI] [PubMed] [Google Scholar]
- 26.Ullah MO, Sweet MJ, Mansell A, Kellie S, Kobe B. 2016. TRIF-dependent TLR signaling, its functions in host defense and inflammation, and its potential as a therapeutic target. J Leukoc Biol 100:27–45. doi: 10.1189/jlb.2RI1115-531R. [DOI] [PubMed] [Google Scholar]
- 27.Gay NJ, Symmons MF, Gangloff M, Bryant CE. 2014. Assembly and localization of Toll-like receptor signalling complexes. Nat Rev Immunol 14:546–558. doi: 10.1038/nri3713. [DOI] [PubMed] [Google Scholar]
- 28.Kawai T, Akira S. 2010. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol 11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 29.Nie L, Cai SY, Shao JZ, Chen J. 2018. Toll-Like receptors, associated biological roles, and signaling networks in non-mammals. Front Immunol 9:1523. doi: 10.3389/fimmu.2018.01523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo L, Lucas RM, Liu L, Stow JL. 2019. Signalling, sorting and scaffolding adaptors for Toll-like receptors. J Cell Sci 133:jcs239194. doi: 10.1242/jcs.239194. [DOI] [PubMed] [Google Scholar]
- 31.Song Y, Shou LM, Ai LY, Bei Y, Chen MT. 2019. The non-immune functions of Toll-like receptors. Crit Rev Eukaryot Gene Expr 29:37–45. doi: 10.1615/CritRevEukaryotGeneExpr.2018027399. [DOI] [PubMed] [Google Scholar]
- 32.Fitzgerald KA, Kagan JC. 2020. Toll-like receptors and the control of immunity. Cell 180:1044–1066. doi: 10.1016/j.cell.2020.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia MM, Goicoechea C, Molina-Alvarez M, Pascual D. 2020. Toll-like receptor 4: a promising crossroads in the diagnosis and treatment of several pathologies. Eur J Pharmacol 874:172975. doi: 10.1016/j.ejphar.2020.172975. [DOI] [PubMed] [Google Scholar]
- 34.Li B, Xia Y, Hu B. 2020. Infection and atherosclerosis: TLR-dependent pathways. Cell Mol Life Sci 77:2751–2769. doi: 10.1007/s00018-020-03453-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohto U. 2017. Conservation and divergence of ligand recognition and signal transduction mechanisms in Toll-like receptors. Chem Pharm Bull (Tokyo) 65:697–705. doi: 10.1248/cpb.c17-00323. [DOI] [PubMed] [Google Scholar]
- 36.Shimizu T. 2017. Structural insights into ligand recognition and regulation of nucleic acid-sensing Toll-like receptors. Curr Opin Struct Biol 47:52–59. doi: 10.1016/j.sbi.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Z, Ohto U, Shimizu T. 2017. Toward a structural understanding of nucleic acid-sensing Toll-like receptors in the innate immune system. FEBS Lett 591:3167–3181. doi: 10.1002/1873-3468.12749. [DOI] [PubMed] [Google Scholar]
- 38.Anwar MA, Shah M, Kim J, Choi S. 2019. Recent clinical trends in Toll-like receptor targeting therapeutics. Med Res Rev 39:1053–1090. doi: 10.1002/med.21553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosadini CV, Kagan JC. 2015. Microbial strategies for antagonizing Toll-like-receptor signal transduction. Curr Opin Immunol 32:61–70. doi: 10.1016/j.coi.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bagheri M, Keller S, Dathe M. 2011. Interaction of W-substituted analogs of cyclo-RRRWFW with bacterial lipopolysaccharides: the role of the aromatic cluster in antimicrobial activity. Antimicrob Agents Chemother 55:788–797. doi: 10.1128/AAC.01098-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khan MM, Ernst O, Sun J, Fraser IDC, Ernst RK, Goodlett DR, Nita-Lazar A. 2018. Mass spectrometry-based structural analysis and systems immunoproteomics strategies for deciphering the host response to endotoxin. J Mol Biol 430:2641–2660. doi: 10.1016/j.jmb.2018.06.032. [DOI] [PubMed] [Google Scholar]
- 42.Mazgaeen L, Gurung P. 2020. Recent advances in lipopolysaccharide recognition systems. Int J Mol Sci 21:379. doi: 10.3390/ijms21020379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ain QU, Batool M, Choi S. 2020. TLR4-targeting therapeutics: structural basis and computer-aided drug discovery approaches. Molecules 25:627. doi: 10.3390/molecules25030627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holdbrook DA, Huber RG, Marzinek JK, Stubbusch A, Schmidtchen A, Bond PJ. 2019. Multiscale modeling of innate immune receptors: endotoxin recognition and regulation by host defense peptides. Pharmacol Res 147:104372. doi: 10.1016/j.phrs.2019.104372. [DOI] [PubMed] [Google Scholar]
- 45.Latty SL, Sakai J, Hopkins L, Verstak B, Paramo T, Berglund NA, Cammarota E, Cicuta P, Gay NJ, Bond PJ, Klenerman D, Bryant CE. 2018. Activation of Toll-like receptors nucleates assembly of the MyDDosome signaling hub. Elife 7:e31377. doi: 10.7554/eLife.31377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nanson JD, Kobe B, Ve T. 2019. Death, TIR, and RHIM: self-assembling domains involved in innate immunity and cell-death signaling. J Leukoc Biol 105:363–375. doi: 10.1002/JLB.MR0318-123R. [DOI] [PubMed] [Google Scholar]
- 47.Ve T, Vajjhala PR, Hedger A, Croll T, DiMaio F, Horsefield S, Yu X, Lavrencic P, Hassan Z, Morgan GP, Mansell A, Mobli M, O’Carroll A, Chauvin B, Gambin Y, Sierecki E, Landsberg MJ, Stacey KJ, Egelman EH, Kobe B. 2017. Structural basis of TIR-domain-assembly formation in MAL- and MyD88-dependent TLR4 signaling. Nat Struct Mol Biol 24:743–751. doi: 10.1038/nsmb.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nanson JD, Rahaman MH, Ve T, Kobe B. 2020. Regulation of signaling by cooperative assembly formation in mammalian innate immunity signalosomes by molecular mimics. Semin Cell Dev Biol 99:96–114. doi: 10.1016/j.semcdb.2018.05.002. [DOI] [PubMed] [Google Scholar]
- 49.De Nardo D, Balka KR, Cardona Gloria Y, Rao VR, Latz E, Masters SL. 2018. Interleukin-1 receptor-associated kinase 4 (IRAK4) plays a dual role in myddosome formation and Toll-like receptor signaling. J Biol Chem 293:15195–15207. doi: 10.1074/jbc.RA118.003314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moncrieffe MC, Bollschweiler D, Li B, Penczek PA, Hopkins L, Bryant CE, Klenerman D, Gay NJ. 2020. MyD88 death-domain oligomerization determines myddosome architecture: implications for Toll-like receptor signaling. Structure 28:281–289.e3. doi: 10.1016/j.str.2020.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang L, Qiao Q, Ferrao R, Shen C, Hatcher JM, Buhrlage SJ, Gray NS, Wu H. 2017. Crystal structure of human IRAK1. Proc Natl Acad Sci U S A 114:13507–13512. doi: 10.1073/pnas.1714386114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kollewe C, Mackensen AC, Neumann D, Knop J, Cao P, Li S, Wesche H, Martin MU. 2004. Sequential autophosphorylation steps in the interleukin-1 receptor-associated kinase-1 regulate its availability as an adapter in interleukin-1 signaling. J Biol Chem 279:5227–5236. doi: 10.1074/jbc.M309251200. [DOI] [PubMed] [Google Scholar]
- 53.Ferrao R, Li J, Bergamin E, Wu H. 2012. Structural insights into the assembly of large oligomeric signalosomes in the Toll-like receptor-interleukin-1 receptor superfamily. Sci Signal 5:re3. doi: 10.1126/scisignal.2003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Napetschnig J, Wu H. 2013. Molecular basis of NF-kappaB signaling. Annu Rev Biophys 42:443–468. doi: 10.1146/annurev-biophys-083012-130338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cohen P, Strickson S. 2017. The role of hybrid ubiquitin chains in the MyD88 and other innate immune signalling pathways. Cell Death Differ 24:1153–1159. doi: 10.1038/cdd.2017.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cohen P, Kelsall IR, Nanda SK, Zhang J. 2020. HOIL-1, an atypical E3 ligase that controls MyD88 signalling by forming ester bonds between ubiquitin and components of the Myddosome. Adv Biol Regul 75:100666. doi: 10.1016/j.jbior.2019.100666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Akutsu M, Dikic I, Bremm A. 2016. Ubiquitin chain diversity at a glance. J Cell Sci 129:875–880. doi: 10.1242/jcs.183954. [DOI] [PubMed] [Google Scholar]
- 58.Lavoie H, Gagnon J, Therrien M. 2020. ERK signalling: a master regulator of cell behaviour, life and fate. Nat Rev Mol Cell Biol 21:607–632. doi: 10.1038/s41580-020-0255-7. [DOI] [PubMed] [Google Scholar]
- 59.Ghosh G, Wang VY, Huang DB, Fusco A. 2012. NF-kappaB regulation: lessons from structures. Immunol Rev 246:36–58. doi: 10.1111/j.1600-065X.2012.01097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Polley S, Passos DO, Huang DB, Mulero MC, Mazumder A, Biswas T, Verma IM, Lyumkis D, Ghosh G. 2016. Structural basis for the activation of IKK1/alpha. Cell Rep 17:1907–1914. doi: 10.1016/j.celrep.2016.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Polley S, Huang DB, Hauenstein AV, Fusco AJ, Zhong X, Vu D, Schrofelbauer B, Kim Y, Hoffmann A, Verma IM, Ghosh G, Huxford T. 2013. A structural basis for IkappaB kinase 2 activation via oligomerization-dependent trans auto-phosphorylation. PLoS Biol 11:e1001581. doi: 10.1371/journal.pbio.1001581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Concetti J, Wilson CL. 2018. NFKB1 and cancer: friend or foe? Cells 7:133. doi: 10.3390/cells7090133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mulero MC, Huxford T, Ghosh G. 2019. NF-kappaB, IkappaB, and IKK: integral components of immune system signaling. Adv Exp Med Biol 1172:207–226. doi: 10.1007/978-981-13-9367-9_10. [DOI] [PubMed] [Google Scholar]
- 64.Tan X, Sun L, Chen J, Chen ZJ. 2018. Detection of microbial infections through innate immune sensing of nucleic acids. Annu Rev Microbiol 72:447–478. doi: 10.1146/annurev-micro-102215-095605. [DOI] [PubMed] [Google Scholar]
- 65.Funami K, Matsumoto M, Oshiumi H, Inagaki F, Seya T. 2017. Functional interfaces between TICAM-2/TRAM and TICAM-1/TRIF in TLR4 signaling. Biochem Soc Trans 45:929–935. doi: 10.1042/BST20160259. [DOI] [PubMed] [Google Scholar]
- 66.Marongiu L, Gornati L, Artuso I, Zanoni I, Granucci F. 2019. Below the surface: the inner lives of TLR4 and TLR9. J Leukoc Biol 106:147–160. doi: 10.1002/JLB.3MIR1218-483RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nimma S, Ve T, Williams SJ, Kobe B. 2017. Towards the structure of the TIR-domain signalosome. Curr Opin Struct Biol 43:122–130. doi: 10.1016/j.sbi.2016.12.014. [DOI] [PubMed] [Google Scholar]
- 68.Zapata JM, Perez-Chacon G, Carr-Baena P, Martinez-Forero I, Azpilikueta A, Otano I, Melero I. 2018. CD137 (4-1BB) signalosome: complexity is a matter of TRAFs. Front Immunol 9:2618. doi: 10.3389/fimmu.2018.02618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, Du F, Ren J, Wu YT, Grishin NV, Chen ZJ. 2015. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347:aaa2630. doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- 70.Dhillon B, Aleithan F, Abdul-Sater Z, Abdul-Sater AA. 2019. The evolving role of TRAFs in mediating inflammatory responses. Front Immunol 10:104. doi: 10.3389/fimmu.2019.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou AY, Shen RR, Kim E, Lock YJ, Xu M, Chen ZJ, Hahn WC. 2013. IKKepsilon-mediated tumorigenesis requires K63-linked polyubiquitination by a cIAP1/cIAP2/TRAF2 E3 ubiquitin ligase complex. Cell Rep 3:724–733. doi: 10.1016/j.celrep.2013.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou R, Zhang Q, Xu P. 2020. TBK1, a central kinase in innate immune sensing of nucleic acids and beyond. Acta Biochim Biophys Sin (Shanghai) 52:757–767. doi: 10.1093/abbs/gmaa051. [DOI] [PubMed] [Google Scholar]
- 73.Fitzgerald KA, Rowe DC, Barnes BJ, Caffrey DR, Visintin A, Latz E, Monks B, Pitha PM, Golenbock DT. 2003. LPS-TLR4 signaling to IRF-3/7 and NF-kappaB involves the toll adapters TRAM and TRIF. J Exp Med 198:1043–1055. doi: 10.1084/jem.20031023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chiang HS, Liu HM. 2018. The molecular basis of viral inhibition of IRF- and STAT-dependent immune responses. Front Immunol 9:3086. doi: 10.3389/fimmu.2018.03086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hayden MS, Ghosh S. 2014. Innate sense of purpose for IKKbeta. Proc Natl Acad Sci U S A 111:17348–17349. doi: 10.1073/pnas.1419689111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu T, Zhang L, Joo D, Sun SC. 2017. NF-kappaB signaling in inflammation. Signal Transduct Target Ther 2:e17023. doi: 10.1038/sigtrans.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mogensen TH. 2018. IRF and STAT transcription factors - from basic biology to roles in infection, protective immunity, and primary immunodeficiencies. Front Immunol 9:3047. doi: 10.3389/fimmu.2018.03047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Peterson LW, Brodsky IE. 2020. To catch a thief: regulated RIPK1 post-translational modifications as a fail-safe system to detect and overcome pathogen subversion of immune signaling. Curr Opin Microbiol 54:111–118. doi: 10.1016/j.mib.2020.01.015. [DOI] [PubMed] [Google Scholar]
- 79.Ke Y, Chen Z, Yang R. 2013. Yersinia pestis: mechanisms of entry into and resistance to the host cell. Front Cell Infect Microbiol 3:106. doi: 10.3389/fcimb.2013.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shi JH, Sun SC. 2018. Tumor necrosis factor receptor-associated factor regulation of nuclear factor kappaB and mitogen-activated protein kinase pathways. Front Immunol 9:1849. doi: 10.3389/fimmu.2018.01849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dumetier B, Zadoroznyj A, Dubrez L. 2020. IAP-mediated protein ubiquitination in regulating cell signaling. Cells 9:1118. doi: 10.3390/cells9051118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wegner KW, Saleh D, Degterev A. 2017. Complex pathologic roles of RIPK1 and RIPK3: moving beyond necroptosis. Trends Pharmacol Sci 38:202–225. doi: 10.1016/j.tips.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Menon MB, Gropengießer J, Ruckdeschel K, Gaestel M. 2018. To die or not to die: regulatory feedback phosphorylation circuits determine receptor-interacting protein kinase-1 (RIPK1) function. Mol Cell Oncol 5:e1396389. doi: 10.1080/23723556.2017.1396389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Feoktistova M, Leverkus M. 2015. Programmed necrosis and necroptosis signalling. FEBS J 282:19–31. doi: 10.1111/febs.13120. [DOI] [PubMed] [Google Scholar]
- 85.Schilling R, Geserick P, Leverkus M. 2014. Characterization of the ripoptosome and its components: implications for anti-inflammatory and cancer therapy. Methods Enzymol 545:83–102. doi: 10.1016/B978-0-12-801430-1.00004-4. [DOI] [PubMed] [Google Scholar]
- 86.Wang Z, Feng J, Yu J, Chen G. 2019. FKBP12 mediates necroptosis by initiating RIPK1-RIPK3-MLKL signal transduction in response to TNF receptor 1 ligation. J Cell Sci 132:jcs227777. doi: 10.1242/jcs.227777. [DOI] [PubMed] [Google Scholar]
- 87.Meng H, Liu Z, Li X, Wang H, Jin T, Wu G, Shan B, Christofferson DE, Qi C, Yu Q, Li Y, Yuan J. 2018. Death-domain dimerization-mediated activation of RIPK1 controls necroptosis and RIPK1-dependent apoptosis. Proc Natl Acad Sci U S A 115:E2001–E2009. doi: 10.1073/pnas.1722013115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Newton K. 2020. Multitasking kinase RIPK1 regulates cell death and inflammation. Cold Spring Harb Perspect Biol 12:a036368. doi: 10.1101/cshperspect.a036368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kesavardhana S, Malireddi RKS, Kanneganti TD. 2020. Caspases in cell death, inflammation, and pyroptosis. Annu Rev Immunol 38:567–595. doi: 10.1146/annurev-immunol-073119-095439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ang RL, Chan M, Ting AT. 2019. Ripoptocide - a spark for inflammation. Front Cell Dev Biol 7:163. doi: 10.3389/fcell.2019.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hopfner KP, Hornung V. 2020. Molecular mechanisms and cellular functions of cGAS-STING signalling. Nat Rev Mol Cell Biol 21:501–521. doi: 10.1038/s41580-020-0244-x. [DOI] [PubMed] [Google Scholar]
- 92.Cheng Z, Dai T, He X, Zhang Z, Xie F, Wang S, Zhang L, Zhou F. 2020. The interactions between cGAS-STING pathway and pathogens. Signal Transduct Target Ther 5:91. doi: 10.1038/s41392-020-0198-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fan X, Jin T. 2019. Structures of RIG-I-like receptors and insights into viral RNA sensing. Adv Exp Med Biol 1172:157–188. doi: 10.1007/978-981-13-9367-9_8. [DOI] [PubMed] [Google Scholar]
- 94.Brisse M, Ly H. 2019. Comparative structure and function analysis of the RIG-I-like receptors: RIG-I and MDA5. Front Immunol 10:1586. doi: 10.3389/fimmu.2019.01586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rehwinkel J, Gack MU. 2020. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat Rev Immunol 20:537–551. doi: 10.1038/s41577-020-0288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Refolo G, Vescovo T, Piacentini M, Fimia GM, Ciccosanti F. 2020. Mitochondrial interactome: a focus on antiviral signaling pathways. Front Cell Dev Biol 8:8. doi: 10.3389/fcell.2020.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ren Z, Ding T, Zuo Z, Xu Z, Deng J, Wei Z. 2020. Regulation of MAVS expression and signaling function in the antiviral innate immune response. Front Immunol 11:1030. doi: 10.3389/fimmu.2020.01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang X, Lin G, Han Z, Chai J. 2019. Structural biology of NOD-like receptors. Adv Exp Med Biol 1172:119–141. doi: 10.1007/978-981-13-9367-9_6. [DOI] [PubMed] [Google Scholar]
- 99.Christgen S, Kanneganti TD. 2020. Inflammasomes and the fine line between defense and disease. Curr Opin Immunol 62:39–44. doi: 10.1016/j.coi.2019.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xue Y, Enosi Tuipulotu D, Tan WH, Kay C, Man SM. 2019. Emerging activators and regulators of inflammasomes and pyroptosis. Trends Immunol 40:1035–1052. doi: 10.1016/j.it.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 101.Hayward JA, Mathur A, Ngo C, Man SM. 2018. Cytosolic recognition of microbes and pathogens: inflammasomes in action. Microbiol Mol Biol Rev 82:e00015-18. doi: 10.1128/MMBR.00015-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lu F, Lan Z, Xin Z, He C, Guo Z, Xia X, Hu T. 2020. Emerging insights into molecular mechanisms underlying pyroptosis and functions of inflammasomes in diseases. J Cell Physiol 235:3207–3221. doi: 10.1002/jcp.29268. [DOI] [PubMed] [Google Scholar]
- 103.Platnich JM, Muruve DA. 2019. NOD-like receptors and inflammasomes: a review of their canonical and non-canonical signaling pathways. Arch Biochem Biophys 670:4–14. doi: 10.1016/j.abb.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 104.Schnappauf O, Chae JJ, Kastner DL, Aksentijevich I. 2019. The pyrin inflammasome in health and disease. Front Immunol 10:1745. doi: 10.3389/fimmu.2019.01745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shen C, Sharif H, Xia S, Wu H. 2019. Structural and mechanistic elucidation of inflammasome signaling by cryo-EM. Curr Opin Struct Biol 58:18–25. doi: 10.1016/j.sbi.2019.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schmidt FI. 2019. From atoms to physiology: what it takes to really understand inflammasomes. J Physiol 597:5335–5348. doi: 10.1113/JP277027. [DOI] [PubMed] [Google Scholar]
- 107.Deets KA, Vance RE. 2021. Inflammasomes and adaptive immune responses. Nat Immunol 22:412–422. doi: 10.1038/s41590-021-00869-6. [DOI] [PubMed] [Google Scholar]
- 108.Zheng D, Liwinski T, Elinav E. 2020. Inflammasome activation and regulation: toward a better understanding of complex mechanisms. Cell Discov 6:36. doi: 10.1038/s41421-020-0167-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lee FF, Davidson K, Harris C, McClendon J, Janssen WJ, Alper S. 2020. NF-kappaB mediates lipopolysaccharide-induced alternative pre-mRNA splicing of MyD88 in mouse macrophages. J Biol Chem 295:6236–6248. doi: 10.1074/jbc.RA119.011495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Manes NP, Nita-Lazar A. 2018. Application of targeted mass spectrometry in bottom-up proteomics for systems biology research. J Proteomics 189:75–90. doi: 10.1016/j.jprot.2018.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Martinez M, Bruce NJ, Romanowska J, Kokh DB, Ozboyaci M, Yu X, Ozturk MA, Richter S, Wade RC. 2015. SDA 7: a modular and parallel implementation of the simulation of diffusional association software. J Comput Chem 36:1631–1645. doi: 10.1002/jcc.23971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Qin S, Pang X, Zhou HX. 2011. Automated prediction of protein association rate constants. Structure 19:1744–1751. doi: 10.1016/j.str.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Angermann BR, Klauschen F, Garcia AD, Prustel T, Zhang F, Germain RN, Meier-Schellersheim M. 2012. Computational modeling of cellular signaling processes embedded into dynamic spatial contexts. Nat Methods 9:283–289. doi: 10.1038/nmeth.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Manes NP, Angermann BR, Koppenol-Raab M, An E, Sjoelund VH, Sun J, Ishii M, Germain RN, Meier-Schellersheim M, Nita-Lazar A. 2015. Targeted proteomics-driven computational modeling of macrophage S1P chemosensing. Mol Cell Proteomics 14:2661–2681. doi: 10.1074/mcp.M115.048918. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Molecular reaction network of core elements of the TLR4 pathway. Instances of multiple edges between nodes indicates that there are multiple molecular interaction mechanisms. For example, the NF-κB proteins (p105, p100, p50, p52, RelA, RelB, and cRel) can dimerize, and each dimer can associate with an IκB protein (IκBa, IκBb, IκBe, p105, p100; the other IκB proteins are not shown). Download FIG S1, PDF file, 1.1 MB (1.1MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Molecular reaction network of core elements of the cytosolic PRR pathways. The MAVS and STING pathways are on the left, and the inflammasomes are on the right. The pathway diagram spans only from the PAMPs to selected major effectors (the TRAFs, the transcription factors, and GSDMD). MAVS is active at the cytosolic side of the outer mitochondrial membrane, and STING is active at the cytosolic side of the ER membrane. Download FIG S2, PDF file, 0.6 MB (660.9KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
(Tab A) Ligands, small molecules, and “System” molecules. (Tab B) Proteoforms. (Tab C) Bimolecular interactions. Download Data Set S1, XLSX file, 0.2 MB (248.5KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.