FIG 1.
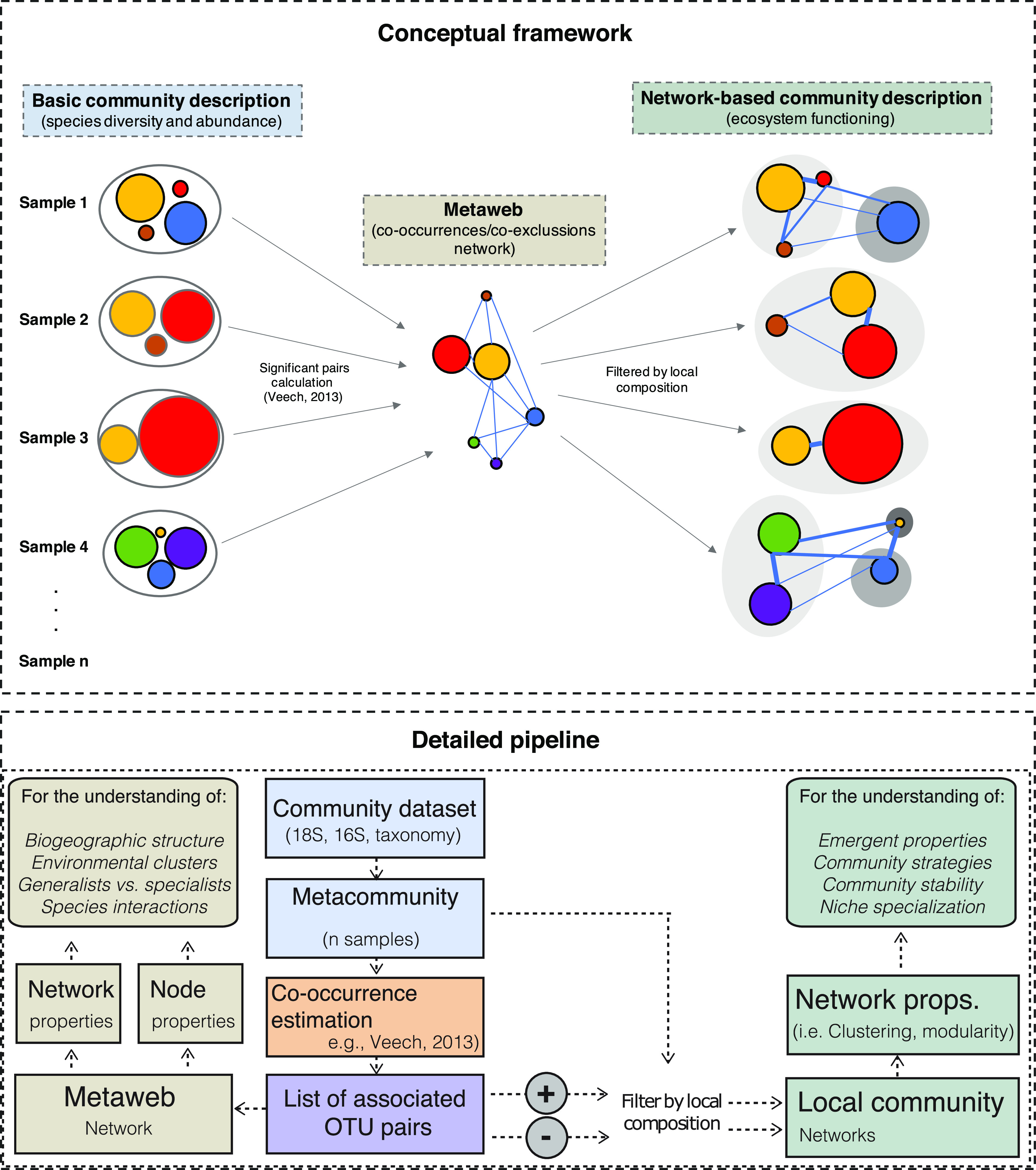
Constructing local community networks from metawebs. Association networks using various spatial scales have been successfully applied to understand the organization and environmental preferences of microbes (15), the fraction of positive trophic interactions (20), generalist versus specialist strategies of particular microbes (81), or even ecological guilds according to potential biotic interactions once the effect of the environment has been removed (Ortiz-Álvarez et al., submitted). This approach is a common strategy in global or regional studies to give general insights of particular metacommunities or larger spatial scales. However, these networks do not inform about the actual arrangements of species in local communities, where species may be loosely or densely connected or display local adaptations to niches or functional guilds. Inferring the local network properties of individual samples characterizes the microbiome of a given sample in terms of its association structure, providing a unique layer of information when studying the biodiversity and stability of a sample or monitoring its evolution in time and during environmental disturbances. Other studies have theorized about how a biogeographical distribution of species interactions is arranged inlocal communities within a given metacommunity or metaweb (82), although this requires the understanding of all the interactions, which is not always possible. Here we combine the metaweb association patterns with local species arrangements to retrieve properties related to particular association arrangements. Considering n local communities within a metaweb, we infer significantly associated pairs of species (both positively and negatively associated). These pairs are later sorted for each local arrangement, so only pairs present in each individual sample are considered, to construct local networks, with particular network properties requiring an ecological interpretation. The ecological interpretation varies according to the positive or negative nature of the associations used to construct the networks, leading to quantifying and understanding the effect of ecological disturbance in ecosystems. Now we define the most important network properties used in our study. Connected components are the subnetwork in which any two nodes connect to each other by edges, that lack connection to any other node in the full network. The clustering coefficient is the measure of the degree to which nodes in a graph tend to cluster together in terms of connected triangles (three nodes that are connected with three edges) in the network (73, 74). The average path length is the mean of the minimal number of required edges to connect any two nodes (73, 74). Modularity is the measure of the strength of a partition into modules (groups of nodes). A good network partition harbors a higher proportion of edges inside modules compared to the proportion of edges between them (72).
