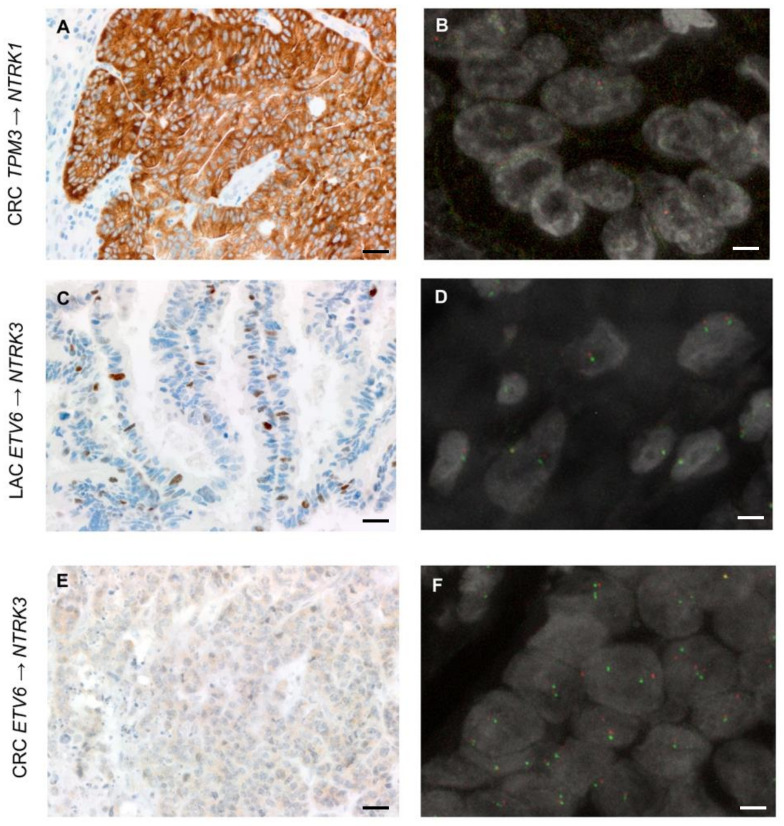
Figure 5.
Representative IHC and FISH images of the cases identified as harbouring an NTRK gene fusion (scale bars—IHC images: 50 μm; scale bars—FISH images = 5 μm). (A,B) The FISH test for the CRC harbouring the intrachromosomal TPM3→NTRK1 rearrangement, as identified by NGS, and displaying strong and diffuse panTRK expression by IHC was suboptimal in terms of quality. Following multiple attempts in all of the available tumour blocks, in scattered tumour cells showing red and green signals, the NTRK1 probes appeared slightly split-apart. This FISH test would have led to an indeterminate result in a diagnostic setting. (C,D) In the LAC sample harbouring the ETV6→NTRK3 fusion, occasional (<10%) nuclei with signals pertaining to NTRK3 probes that were slightly split-apart could be identified. It should be acknowledged that according to standard FISH scoring criteria, (i) the distance between signals may be debatable and (ii) the test would be considered negative, given that the positive nuclei do not reach the 15% cut-off used in the diagnostic setting. (E,F) The CRC sample harbouring the ETV6→NTRK3 fusion showed split-apart NTRK3 signals in about 17% of the tumour cell population (here illustrated). This FISH test meets the criteria for the identification of a genetic rearrangement involving NTRK3.