Figure 1.
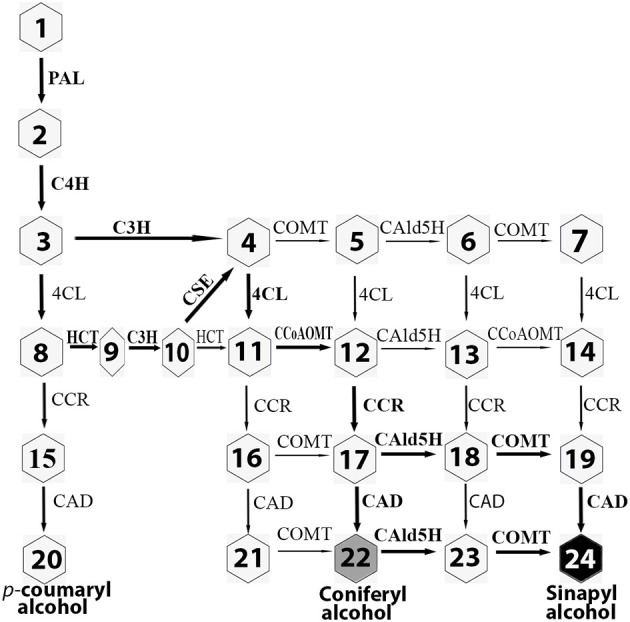
Monolignol biosynthetic pathway in poplar. Bold arrows indicate the main metabolic flux. Eleven enzyme families are PAL, phenylalanine ammonia-lyase; C4H, cinnamate 4-hydroxylase; C3H, p-coumaroyl-CoA 3-hydroxylase; 4CL, p-coumarate CoA ligase; HCT, hydroxycinnamoyltransferase; CSE, caffeoyl shikimate esterase; CCoAOMT, caffeoyl-CoA O-methyltransferase; CCR, cinnamoyl-CoA reductase; CAld5H, coniferaldehyde 5-hydroxylase; COMT, caffeic acid 3-O-methyltransferase; CAD, cinnamyl alcohol dehydrogenase. The metabolites are shown in numbers. 1: phenylalanine; 2: cinnamic acid; 3: 4-hydroxycinnamic acid; 4: caffeic acid; 5: ferulic acid; 6: 5-hydroxyferulic acid; 7: sinapic acid; 8: p-coumaroyl-CoA; 9: p-coumaroyl shikimic acid; 10: caffeoyl shikimic acid; 11: caffeoyl-CoA; 12: feruloyl-CoA; 13: 5-dydroxyferuloyl-CoA; 14: sinapoyl-CoA; 15: p-coumaraldehyde; 16: caffeyl aldehyde; 17: coniferaldehyde; 18: 5-hydroxyconiferaldehyde; 19: sinapaldehyde; 20: p-coumaryl alcohol (H monolignol); 21: caffeyl alcohol; 22: coniferyl alcohol (G monolignol); 23:5-hydroxyconiferyl alcohol; 24: sinapyl alcohol (S monolignol).
