Figure 1 |. CRISPR screens identify MPP8 as a vulnerability for myeloid leukemia.
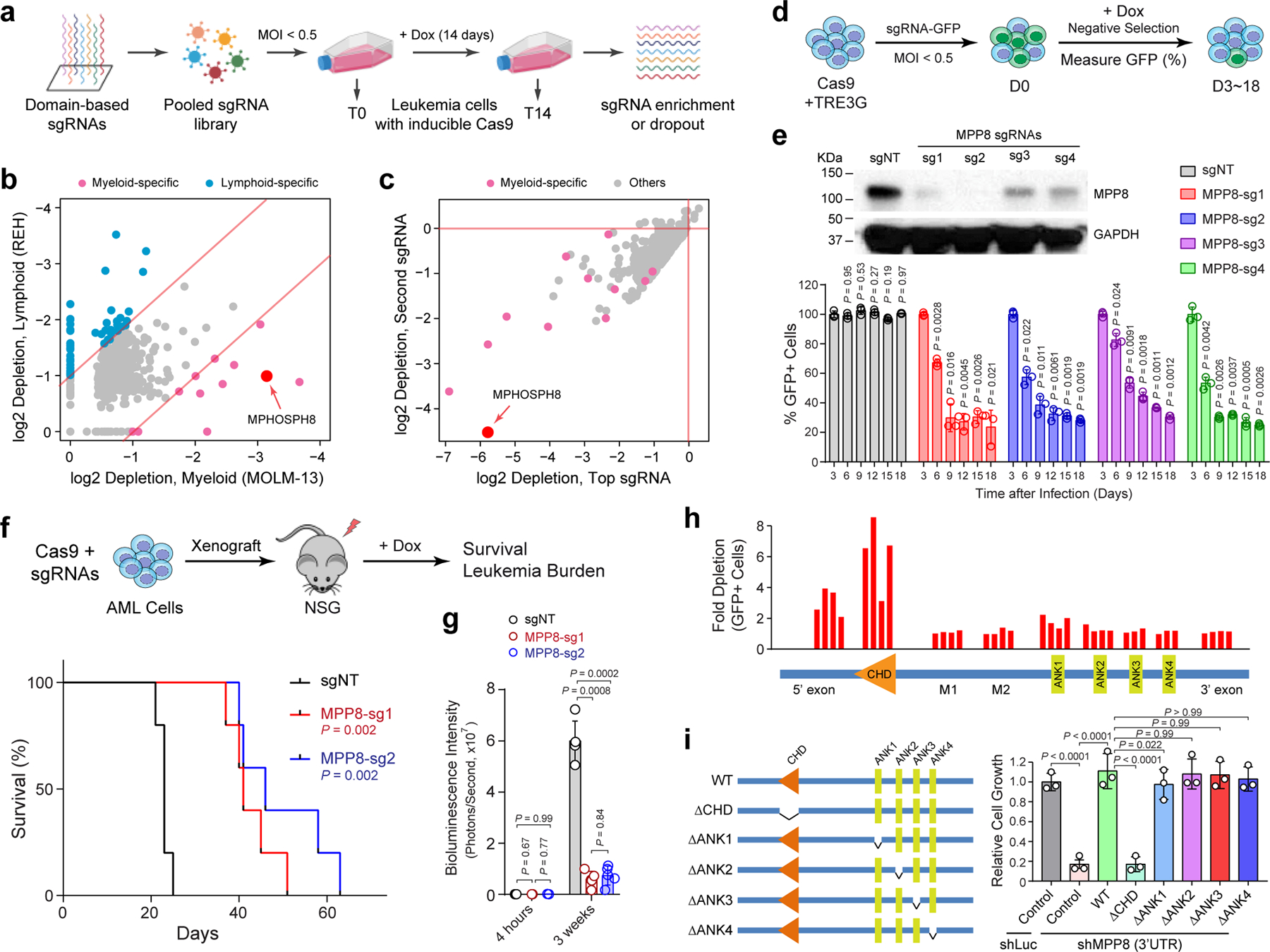
a, Schematic of the domain-focused CRISPR dependency screen. b, Scatterplot depicting the dependency genes identified by CRISPR screens in AML (MOLM-13) and ALL (REH) cells. Red and blue dots indicate myeloid- and lymphoid-specific dependency genes, respectively. x- and y-axis indicate the log2 fold depletion of sgRNA abundance between day 0 and day 14, respectively. Results are based on mean values of all sgRNAs from two independent screens. c, Scatterplot depicting the myeloid-specific dependency genes (red dots) by the top and second dropout sgRNAs between day 0 and day 14 in MOLM-13 cells. d, Schematic of the negative-selection competition assay. e, MPP8 depletion by independent sgRNAs impaired MOLM-13 cell growth relative to the non-targeting control sgRNA (sgNT). Results are mean ± SD and analyzed by a two-way ANOVA with Dunnett’s multiple comparison test. MPP8 depletion was confirmed by Western blot. f, Kaplan-Meier survival curves of NSG mice xenografted with control (sgNT) or MPP8-deficient (MPP-sg1 and MPP8-sg2) MOLM-13 cells. P values by a log-rank Mantel-Cox test. g, Quantification of leukemia burden by bioluminescent imaging 4 hours (baseline) and 3 weeks post-xenograft. Results are mean ± SD (N = 5 mice per group) and analyzed by a two-way ANOVA with Tukey’s multiple comparison test. h, CRISPR-based mutagenesis of MPP8 domains by the negative-selection competition assay in MOLM-13 cells. i, Re-introduction of WT but not CHD-deleted MPP8 rescued the cell growth defects of MPP8-depleted MOLM-13 cells. Results are mean ± SD (N = 3 independent experiments) and analyzed by a two-way ANOVA with Tukey’s test.
