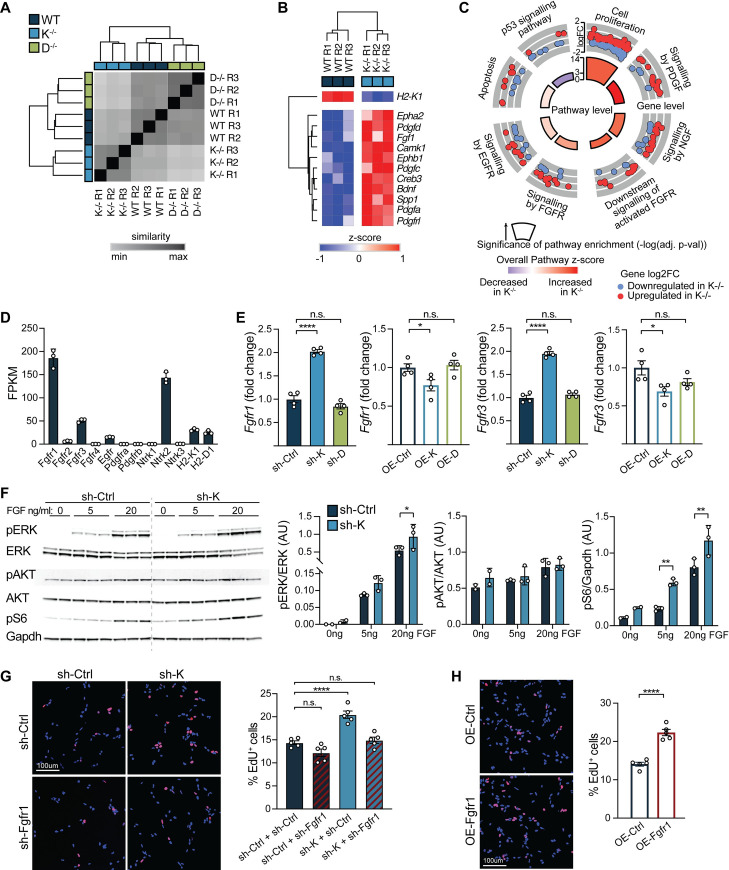
Fig 3. H2-Kb negatively regulates NSPC proliferation by inhibiting Fgfr signaling.
RNA-seq analysis of primary hippocampal WT, K−/−, and D−/− NSPCs cultured under self-renewal conditions. Pairwise distances (Euclidean distance) between RNA-seq libraries prepared from WT, K−/−, and D−/− primary hippocampal NSPCs. Dendrogram was calculated by unsupervised hierarchical clustering (average linkage). (n = 3 replicates/group [denoted R1–R3]) (A). (B) Unsupervised hierarchical clustering of growth factor signaling-related genes identified to be differentially expressed between WT and K−/− hippocampal NSPCs using RNA-seq analysis (z-score normalized, rows). (C) Cellular and signaling pathways identified by pathway enrichment analysis (ConsensusPathDB) of genes differentially expressed between WT and K−/− NSPCs are subdivided in gray. Outer ring illustrates expression of altered genes (log2 fold change between K−/− vs WT) within each pathway. Inner ring illustrates significance (−log10 adj. p-value) and overall expression levels of genes within each pathway (z-score). (D) Expression (FPKM) of growth factor receptors and H2-K1 and H2-D1 was evaluated in WT NSPCs using RNA-seq. n = 3 replicates/group for all RNA-seq analysis. (E) WT adult NSPCs were infected with either sh-K and sh-control (sh-Ctrl) or Nestin-driven K overexpression (OE-K) and GFP control (OE-Ctrl) lentiviruses. Fgfr1/Fgfr3 expression were measured using quantitative RT-PCR 72 hours after infection. n = 4 replicates/group. (F) Representative western blot (left panel) and quantifications (right panels) of activated Akt, Erk, and ribosomal protein S6 in adult WT NSPCs infected with lentiviruses encoding sh-K or sh-control (sh-Ctrl). NSPCs were grown in the absence of (0 ng/mL), low (5 ng/mL), or regular (20 ng/mL) FGF level for 2 hours. n = 2–3 replicates/time point/group; data represented as mean ± SD. (G) Representative images (left panel) and quantification (right panel) of adult NSPCs infected with sh-K or sh-Ctrl in combination with lentiviruses expressing shRNAs against Fgfr1 (sh-Fgfr1) or control (sh-Ctrl). NSPCs were grown under self-renewal conditions and treated with EdU for 6 hours to test for proliferation. Percentage of EdU+ cells are shown. n = 5 replicates/group. (H) Representative images (left panel) and quantification (right panel) of WT adult NSPCs infected with Nestin-Fgfr1 overexpression (OE-Fgfr1) or GFP control (OE-Ctrl) lentiviruses. NSPCs and cultured under self-renewal conditions treated with EdU for 6 hours to test for proliferation. Percentage of EdU+ cells are shown. n = 5 replicates/group. Data represented as mean ± SEM. One-way ANOVA with Dunnett’s multiple comparisons test (E, G), two-way ANOVA with Sidak’s multiple comparisons test (F), and Student t test (H). *p < 0.05, **p < 0.01, ****p < 0.0001. Data used to generate this figure can be found in the Supporting information Excel spreadsheet (S1 Data). EdU, 5-ethynyl-2′-deoxyuridine; EGFR, epidermal growth factor receptor; FGF, fibroblast growth factor; Fgfr, fibroblast growth factor receptor; Fgfr1, fibroblast growth factor receptor 1; Gapdh, glyceraldehyde 3-phosphate dehydrogenase; GFP, green fluorescent protein; NGF, nerve growth factor; n.s., not significant; NSPC, neural stem and progenitor cell; PDGF, platelet-derived growth factor; RNA-seq, RNA-sequencing; RT-PCR, reverse transcription polymerase chain reaction; shRNA, short hairpin RNA; WT, wild-type.