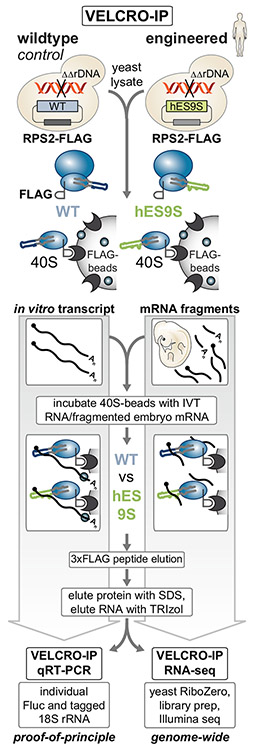
Figure 2. Development of VELCRO-IP RNA-seq to identify global ES-mRNA interactions.
Schematic representation of the VELCRO-IP approach. Yeast strains expressing chimeric (hES9S) orWT ribosomes are generated by rDNA complementation. The same strains also carry endogenously C-terminally FLAG-tagged RPS2/uS5. 40S ribosomal subunits from powderized lysates of each strain are isolated on FLAG agarose beads and washed. For VELCRO-IP qRT-PCR (proof of principle), in vitro transcripts (IVTs) (see Figure 3) are incubated with ribosome beads. Upon 3xFLAG peptide elution of 40S-RNA complexes, total RNA is eluted, and IVT RNA enrichment is determined by qRT-PCR specific for Fluc and the 18S rRNA tag. For VELCRO-IP RNA-seq (genome-wide), mRNAs from total RNA from stage E11.5 mouse embryos are purified and fragmented to 100–200 nt, and refolded RNA fragments are used as input for IP and FLAG elution of mRNA-ribosome complexes. After yeast rRNA depletion from eluted RNAs, ribosome-bound mRNA fragments are sequenced to identify hES9S-specific mouse mRNA elements.