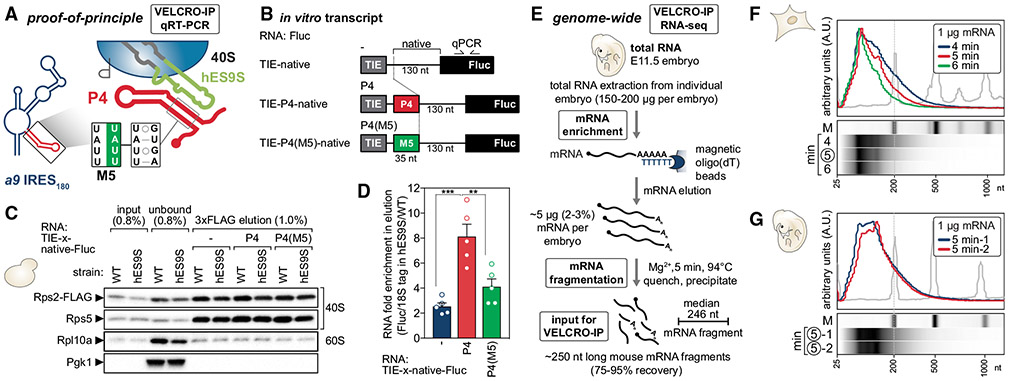
Figure 3. VELCRO-IP qRT-PCR serves as a proof of principle and mouse embryo mRNA fragmentation.
(A) VELCRO-IP qRT-PCR: a zoomed-in view on the interactions between hES9S and Hoxa9 P4 stem-loop (Leppek et al., 2020) or other target 5′ UTRs that can be identified by VELCRO-IP. The 4-nt inactive P4 mutant M5 (P4(M5)) serves as a negative control.
(B) IVTs of 475–510 nt in length contain the native spacer (–, negative control), P4-native (P4), or P4(M5)-native (P4(M5)) embedded in flanking constant regions (5′ TIE and 3′ Fluc ORF sequence) (see Leppek et al., 2020). The Fluc ORF portion can be used for qPCR amplification to compare the three RNA constructs. TIE, translation inhibitory element.
(C) Western blot (WB) analysis of same volumes of lysate (input), unbound fraction, and 3xFLAG peptide-eluted protein from beads to monitor ribosome enrichment of tagged (Rps2-FLAG) and untagged (Rps5) 40S and 60S (Rpl10a) components in IVT RNA samples, in combination with WT and hES9S yeast ribosomes. Cytoplasmic enzyme Pgk1 served as a negative control. The fraction loaded of input, unbound, and elution samples is expressed as a percentage of the original lysate volume. A representative experiment of n = 5 is shown.
(D) Analysis of total RNA in the 3xFLAG peptide elution by qRT-PCR using the same volumes of RNA per sample for the RT. Fluc transcript enrichment was assessed by normalizing Ct values to those of the respective 18S rRNA tag to control for ribosome-IP efficiency per sample. Respective hES9S samples were compared with WT samples to assess RNA fold enrichment of IVT RNAs. Average RNA fold enrichment ± SEM, n = 5. See also Figures S2E-S2G.
(E) Schematic of embryo mRNA fragmentation for VELCRO-IP RNA-seq. Total RNA extraction of stage E11.5 mouse embryos yields 2%–3% of mRNA isolated on oligo(dT) beads. mRNA is fragmented with magnesium ions to a length of 100–200 nt, which overall recovers >75% of input mRNAs as fragments.
(F) Fragmented mouse mRNAs from C3H10T1/2 cells in 1-μg aliquots at different time points of fragmentation (4, 5, and 6 min) were analyzed on an mRNA Pico Chip (Agilent) on a Bioanalyzer (Agilent). A zoomed-in view of the Bioanalyzer quantification (top) and virtual gel images (bottom) is shown. The marker (gray line, lane M) is overlaid for reference. See also Figures S3A-S3C.
(G) Fragmented mouse mRNAs from stage E11.5 embryos in 1-μg aliquots fragmented for 5 min at 94°C from two independent repeats of embryo harvest, RNA isolation, mRNA purification, and fragmentation (1 and 2). This yields fragments of 100–200 nt. RNAs were analyzed as in (F). See also Figure S3C.