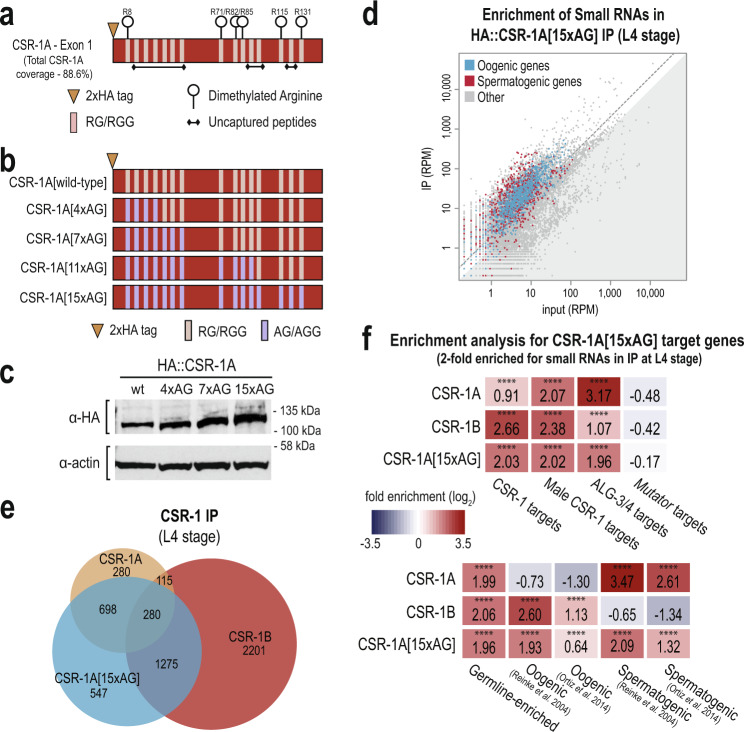
Fig. 6. Methylation of CSR-1A N-terminal exon promotes binding preference for spermatogenic siRNAs.
a Graphical display of dimethylation modifications detected on CSR-1A by mass spectrometry following IP. b Schematic representation of the series of arginine to alanine mutations created by CRISPR to generate the unmethylatable CSR-1A[15xAG]. c Western blot for HA::CSR-1A in the RG-to-AG mutants. Actin is shown as a loading control. The blot has been reproduced. d Normalized reads for spermatogenic and oogenic genes (defined by Reinke et al., 2004) from HA::CSR-1A[15xAG] IP compared to input. Spermatogenic and oogenic genes are highlighted in red and blue, respectively. Gray dotted line indicates twofold enrichment in IP relative to input. e Venn diagram indicates overlap of CSR-1A, CSR-1B, and CSR-1A[15xAG] target genes at L4 stage. f Enrichment analysis (log2(fold enrichment)) examining the overlap of CSR-1A, CSR-1B, and CSR-1A[15xAG] target genes with known targets of the CSR-1, male CSR-1, ALG-3/4, and mutator small RNA pathways and oogenesis and spermatogenesis-enriched genes. See Materials and Methods for gene list information. Two-tailed p values for enrichment was calculated using the Fisher’s exact test function in R. n.s denotes not significant and indicates a p value >0.05 and **** indicates a p value ≤0.0001. See Supplementary Data 7 for more details regarding statistical analysis. Source data are provided as a Source data file.