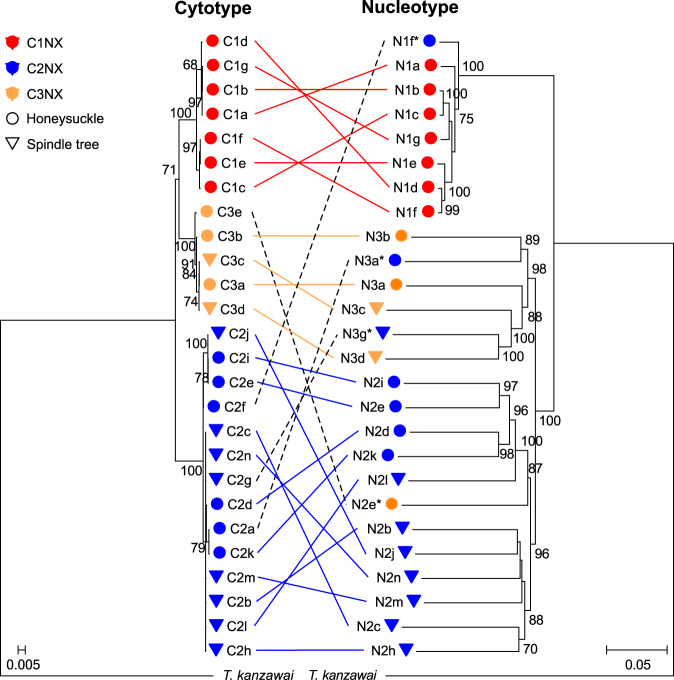
Fig. 2. Mitochondrial and nuclear genome phylogenies of field-derived Tetranychus urticae iso-female lines.
At left is a maximum likelihood analysis based on the optimal partitioning scheme of 13 protein-coding mitochondrial genes. At right is a neighbour-joining analysis using whole nuclear genomes. In both analyses, Tetranychus kanzawai is included as an outgroup. The scale bar of the mitochondrial phylogeny represents the nucleotide substitutions per site, the scale bar of the nuclear genome phylogeny represents substitutions per nuclear SNP position. Mitochondrial and nuclear phylogenies for the same field-derived lines are labelled and connected by lines, with colours according to their cytotype and shapes according to their native hosts. Cyto-nuclear hybrids are connected with dotted black lines and have asterisks next to their nuclear name. Only bootstraps values ≥65 are shown.