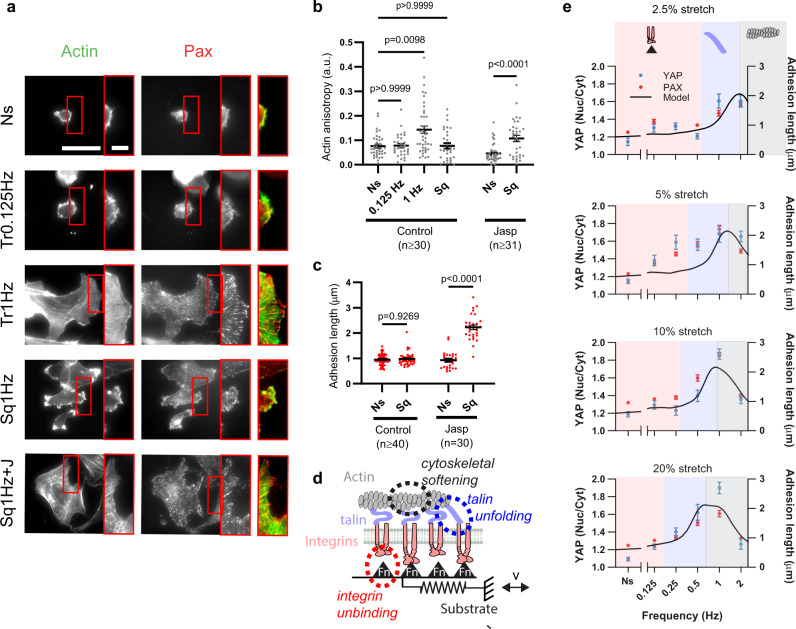
Fig. 2. A molecular clutch model considering mechanosensing and cytoskeletal softening predicts the response to stretch.
a Actin and paxillin stainings for cells either not stretched or stretched by 10% with triangular signals (0.125 Hz, 1 Hz) or square signals (Sq, 1 Hz, with or without Jasplakinolide treatment). Areas circled in red are shown magnified at the right of each image, and shown as a merged image (actin, green, paxillin, red). Scale bar is 50 µm, and 10 µm in magnifications. b, c corresponding quantifications of actin anisotropy (b) and adhesion length (c) for control cells, and cells treated with Jasplakinolide. n numbers are cells. Statistical significance was assessed with two-way ANOVA. d Cartoon of computational clutch model (see “Methods” section). The model considers a relative speed of movement v between cell and substrate (given by stretch). The substrate is represented by an elastic spring with binding sites to integrins (via fibronectin, Fn triangles), which in turn connect to talin and actin. As stretch applies forces, these can lead to integrin unbinding, talin unfolding, or cytoskeletal softening. e Model predictions (black line) overlaid on experimental YAP and paxillin results from Fig. 1. The only parameter changing between simulations is stretch amplitude (1.5, 2, 2.5, and 3 µm for 2.5, 5, 10, and 20% stretch). Areas shaded in pink, blue, and gray show the regions dominated respectively by integrin unbinding, talin unfolding, and cytoskeletal softening. Data are shown as mean ± s.e.m.