Graphical abstract
Keywords: Protein-protein Interaction, Co-evolutionary pairings, Intra-protein co-evolution, Interprotein co-evolution, Mutual Information
Highlights
-
•
Co-Var identifies co-evolutionary pairings in proteins and protein–protein interactions.
-
•
High degree co-evolutionary pairings occur in interface and non-interface regions.
-
•
Co-Var web-server allows the prediction of functionally important residue positions.
Abstract
Proteins involved in interactions throughout the course of evolution tend to co-evolve and compensatory changes may occur in interacting proteins to maintain or refine such interactions. However, certain residue pair alterations may prove to be detrimental for functional interactions. Hence, determining co-evolutionary pairings that could be structurally or functionally relevant for maintaining the conservation of an inter-protein interaction is important. Inter-protein co-evolution analysis in several complexes utilizing multiple existing methodologies suggested that co-evolutionary pairings can occur in spatially proximal and distant regions in inter-protein interactions. Subsequently, the Co-Var (Correlated Variation) method based on mutual information and Bhattacharyya coefficient was developed, validated, and found to perform relatively better than CAPS and EV-complex. Interestingly, while applying the Co-Var measure and EV-complex program on a set of protein–protein interaction complexes, co-evolutionary pairings were obtained in interface and non-interface regions in protein complexes. The Co-Var approach involves determining high degree co-evolutionary pairings that include multiple co-evolutionary connections between particular co-evolved residue positions in one protein with multiple residue positions in the binding partner. Detailed analyses of high degree co-evolutionary pairings in protein–protein complexes involved in cancer metastasis suggested that most of the residue positions forming such co-evolutionary connections mainly occurred within functional domains of constituent proteins and substitution mutations were also common among these positions. The physiological relevance of these predictions suggested that Co-Var can predict residues that could be crucial for preserving functional protein–protein interactions. Finally, Co-Var web server (http://www.hpppi.iicb.res.in/ishi/covar/index.html) that implements this methodology identifies co-evolutionary pairings in intra and inter-protein interactions.
1. Introduction
Intra-protein co-evolution which involves compensatory substitutions within proteins can restore functionality by sustaining the fitness of the protein under constraints imposed by physico-chemical interaction forces, structural and folding associated factors [1], [2], [3], [4]. Multiple approaches have been utilized to study intra-molecular co-evolution such as substitution pattern correlations, mutual information of amino acid frequencies between positions in a multiple sequence alignment (MSA), analysis of evolutionary phylogenetic trees etc. [5], [6], [7], [8]. Further, a number of coevolution-based contact prediction methods which include adjustments for direct and indirect couplings have been developed for monomeric proteins [9], [10], [11], [12], [13]. In general, it has been observed that interacting residues in proximity tend to co-evolve [14], [15], [16], [17], [18], [62]. However, large number of coevolutionary connections may occur at less variable positions within a protein family [19]. Moreover, clusters of positions which are usually not in contact but tend to be located near binding regions or active sites have been found to co-evolve [20], [21], [22].
Recently, methods have been developed to predict networks of residues or patterns of co-evolved amino acids within a protein (residue communities). These residues are relevant to protein function, such as protein specificity in protein–protein interactions, phospholipid-binding activity, or conformational changes [23]. Thus, it is possible that coevolution between distant sites can arise from residue–residue interactions due to allosteric interaction networks [24], negative design [25], or codon effects [26] etc. Additionally, a recent report suggests that spatially distant mutational hotspots tend to co-evolve [27], [63]. Moreover, direct couplings between residues distantly located in protein structures (>5 Å and 15 Å apart) have also been identified [28]. Therefore, in addition to direct contacts, allosteric networks between residues at long distances can also be involved in residue–residue couplingSalinas and Ranganathan [29].
Protein-protein interactions are inherently important in signal transduction pathways or metabolic reactions within cells to carry out diverse physiological processes. Single protein molecules may interact fleetingly in transient complexes involved in different types of interactions (for e.g., signaling–effector, enzyme–inhibitor, enzyme–substrate, hormone–receptor etc.) whereas some proteins may exist as parts of multi-subunit enzymes in permanent obligate interactions [30]. During such interactions, correlative sequence evolution can occur between proteins that physically interact or have a functional association in a manner such that amino acid changes at one site in a molecule may give rise to changes in selection pressure at another site in the binding partner. This evolutionary interaction between protein sites in different molecules that undergo compensatory changes to maintain the stability or functions of the interaction over the course of evolution is referred to as inter-protein co-evolution [31]. The observation that interface positions exhibit changes in a correlated manner among interacting molecules lead to the development of methods for predicting contacting pairs of residues from sequence information [32], [33], [34]. Analysis of co-evolution in inter-protein complexes has demonstrated that residues at the interfaces of obligate complexes co-evolve with their interacting partners whereas transient protein interaction complexes have an increased rate of substitution at the interface residues with less correlated mutations occurring across the interface [30]. In general, while functionally important co-evolving residues having high mutual information (MI) occur in structural proximity [35], [36], a fraction of coevolving residue pairs predicted based on direct couplings have recently been shown to occur at distant regions in protein structures [28]. Evolutionary pressure is likely to maintain an interaction between protein interfaces wherein selection restricts amino acid replacements or preserves a degree of conservation in the binding interfaces to maintain functionality of such interactions [31]. In this respect, analysis of molecular co-evolution in inter-protein complexes may be useful for determining co-evolutionary pairings among interface residues and it is likely that coordinated changes at these residue positions are likely to be crucial for a functional interaction between these sets of proteins. In this study, we have developed a method named Co-Var (Correlated Variation) aiming to determine both inter-protein and intra-protein residues that are likely to carry out crucial structural or functional roles in protein–protein interactions via establishing co-evolutionary pairings between themselves. Herein, we have determined the applicability of the Co-Var measure in studying inter-protein co-evolution and compared the methodology with selected protein–protein co-evolution analysis methodologies such as CAPS [37], [21] and EV-complex [33]. However, we observed that co-evolving residues in inter-protein interaction complexes were found to occur in interface regions (close spatial proximity) as well as in non-interface regions. This observation was like a previous study wherein physically-coupled amino-acids at short range distances, and uncoupled amino-acids at long-range distances co-evolve with high mutual information content, however, the signal is stronger for coupled residues [38].
Based on this observation, we have considered the hypothesis that co-evolutionary pairings that occur in interface and non-interface regions could be crucial for native interactions and absence of coordinated changes at these positions are likely to contribute to altered interaction profiles and aberrant complex functionality. Therefore, to study the likely structural or functional relationship between co-evolutionary pairings in interface or non-interface regions and aberrant complex functionality certain protein–protein interaction complexes that exhibit frequent mutations in cancer have been considered. Studies on distribution pattern of disease associated variants have identified that disease causing mis-sense mutations frequently occur at the core region of protein–protein interaction interfaces or at the ligand-binding sites and residues involved in enzymatic function [39], [40]. Thus, co-evolution in certain ligand-receptor proteins as case studies has been studied with the help of Co-Var to exemplify the physiological relevance of predicted co-evolutionary pairings. Therefore, in this work we have implemented the Co-Var methodology, determined its applicability in studying inter-protein co-evolution and utilized it to study inter-cellular interaction complexes involved in cancer metastasis. The composite co-evolutionary measures along with various options to visualize such co-evolutionary pairings onto the representative structure of the complexes have been implemented within Co-Var web server which is freely accessible at http://www.hpppi.iicb.res.in/ishi/covar/index.html.
2. Materials and methods
2.1. Determining co-evolutionary pairs in positive and negative protein–protein interaction complexes
A set of protein–protein interaction complexes (100) were collected from previous published data [41], [42], [43] and complexes satisfying our selection criteria of sufficient number of homologs, availability of crystal structure etc. were considered during this analysis. From this set, 50 protein complexes were selected as the set of interacting complexes that are likely to co-evolve (“positive set”) (Supplementary data 2 Table S1). Additionally, proteins which were known to be non-interacting based on experimental analysis were randomly selected from the Negatome database [44] as the “negative set” (Supplementary data 2 Table S1). The compiled positive and negative set comprising of 50 heterodimeric protein pairs each were subjected to MirrorTree [45], CAPS [37], [21] and EV-complex [33] methods respectively. Other methods such as DCA or GREMLIN which are mainly used for contact prediction have not been considered here since we wanted to capture co-evolved positions in interface and non-interface regions to determine the possible implications of the same. Close orthologs or similar sequences were determined using DELTA-BLAST (Domain enhanced lookup time accelerated BLAST) [46] and taxonomy filtered non-redundant sequences having E-value <= 1E-04, query coverage >= 70%, sequence identity >= 45% were utilized for preparing multiple sequence alignments (MSA) representative of each sequence family with the help of MAFFT [47]. MirrorTree was utilized to determine whether the proteins co-evolve considering alignments of homologous sequences of the representative interacting and non-interacting proteins in each set. Here, tree similarities are quantified with the help of linear correlation by extracting inter-ortholog distance matrices from the MSA-derived trees of orthologous protein sequences in the two families [45]. CAPS was run with the help of default parameters on the set of alignments generated to identify amino acid co-variation with the help of BLOSUM corrected amino acid distances and phylogenetic sequence relationships [37]. EV-complex was utilized to predict inter-evolutionary couplings in the alignment of concatenated sequences (generated internally during the calculation) using a global probability model of sequence co-evolution (pseudo-likelihood maximization) [33]. Subsequently, a distance distribution plot was prepared to analyze the inter-residue distances between the predicted co-evolving residue positions among the interacting proteins (positive set) obtained from CAPS and EV-complex.
2.2. Identifying intra-protein and inter-protein co-evolution with an information theory-based measure (Co-Var)
In information theory, mutual information represents the entropy-based formulation for quantifying the interdependence between the values of two random categorical variables which in this case could be position-wise amino acid frequency distributions [5]. Further, mutual information is defined as the amount of information one variable or amino acid frequency distribution (column A) can tell us about the value of another variable or amino acid frequency distribution (column B). In other words, it is the reduction in uncertainty (entropy) in the value of ‘A,’ if we know the value of ‘B’ and is calculated considering the sum over all the possible combinations of di-residue frequencies [48]. Mutual information (MI) between two aligned columns A and B is calculated as:
wherein, ‘p(a)’ is the frequency of occurrence of each residue in the column ‘A,’ ‘p(b)’ is the frequency of occurrence of each residue in the column ‘B’ and ‘p(a, b)’ represents the di-residue frequency. Additionally, the Bhattacharyya coefficient quantifies the overlap between set of amino acids between a pair of columns. It is a measure of similarity between two datasets or distributions and is used to calculate the amount of overlap between two distributions, by splitting the samples into several partitions.
wherein, BHC(A, B) denotes Bhattacharyya coefficient between positions A and B, ‘p(a)’ and ‘p(b)’ are the amino acid residue frequencies present in the respective positions.
A score is computed considering homologous set of sequences within a protein family to derive intra-residue correlations in a single protein wherein each alignment position is compared to all other positions within the alignment for the protein family under consideration. Alternately, correlated positions between pairs of interacting proteins can also be identified based on the Co-Var score considering the position-wise amino acid frequencies in the multiple sequence alignments of the proteins involved in inter-protein interaction. Correlations between evolutionary patterns within proteins or between proteins may be determined based on the Co-Var score as outlined below:
wherein, Co-Var score(A, B) represents the co-variation score between position A and B, MI (A, B) and BHC(A, B) denote mutual information and Bhattacharyya coefficient between positions A and B. Additionally, in case of intra-protein co-evolution ‘A’ and ‘B’ represent different positions within the multiple sequence alignment of a protein family whereas in inter-protein co-evolution analysis ‘A’ and ‘B’ represent a position in the alignment for the first protein family and another position in the second protein family respectively. The Co-Var methodology to study intra-protein and inter-protein co-evolution has been depicted in Fig. 1 and Fig. 2, respectively.
Fig. 1.
Co-Var methodology for studying intra-molecular co-variation in proteins.
Fig. 2.
Co-Var methodology for studying protein–protein co-evolution.
Close orthologs or similar sequences were determined using DELTA-BLAST (Domain enhanced lookup time accelerated BLAST) [46] and minimum 50 sequences were considered for generating an MSA representative of each family wherein the first sequence in each alignment was considered as the reference sequence. While detecting co-evolution, alignment shuffling was performed with a view to reduce the influence of phylogenetic relationships. Shuffling was performed by randomly selecting orthologous sequences in each family such that amino acids across the column exhibited variation. Further, multiple instances of the program were run and co-evolutionary pairings that are consistently identified across the different runs based on z-score threshold were considered such that additional statistical significance can be assigned to the co-evolving positions. After the calculation of the Co-Var scores, the residue pairs and scores were mapped to a corresponding reference sequence and structure for each set and average Co-Var score and corresponding z-scores across the runs were determined. While it is difficult to classify the residue positions as true negative or false positives without extensive experimental analysis of the predicted co-evolving positions; in general, based on previous works it has been observed that interface residues tend to co-evolve. Interface pairs were calculated using PISA (available in ccp4mg version 2.8.1); [49]. Considering interface pairs as true positive co-evolving set, a ROC analysis was performed for 25 complexes in the positive set. Herein, it was considered essential to achieve sensitivity and specificity values of around 0.7 while reducing the FDR as far as practical. At higher negative z-scores than −1 the sensitivity and specificity fall drastically to lower than 0.3 in all cases (data not shown). Herein, it was observed that Co-Var scores corresponding to lower (negative) z-scores are indicative of higher likelihood of co-variation. Based on this analysis, we selected z <=-1 (Z-score was calculated for each co-evolutionary pairing based on the Co-Var scores) as the threshold for our analysis. Thus, co-evolving positions having significant Co-Var score and z-scores which are reported in multiple runs (5) of the program have been considered for further analysis. This is because the residue pairing positions that consistently received a significant score, when considering different alignments (due to sequence shuffling) have a higher likelihood of being co-evolved.
2.3. Benchmarking of predicted co-evolutionary positions in protein–protein interaction complexes
In order to determine the applicability of Co-Var methodology in studying inter-protein co-evolution the ‘percentage of co-evolved pairs’ in interacting (positive) and non-interacting (negative) proteins has been considered as an index. In this study, it was observed that multiple methods were able to detect spatially distant co-evolving positions and since we wanted to explore this observation in detail, we have used an index other than the area under the curve for performance comparison. The ‘percentage of co-evolved pairs’ predicted for each positive and negative set pair by CAPS and EV-complex was also determined for these complexes. Based on these analyses, we have determined whether these indices can successfully differentiate between the positive and negative set. Subsequently, by considering a reference structure for each of the positive set of complexes, a distance distribution plot was prepared to analyze the inter-residue distances between the Co-Var predicted co-evolving residues. Additionally, two measures that capture the co-evolving pairs in the overall complex [Percentage of co-evolved pair that occur at interface (IC)] and those that occur at the interface [Percentage of interacting pair that are co-evolved (PC)] were also computed. Here, co-evolved pairs predicted by Co-Var and EV-complex for the positive set were considered. For this purpose, co-evolving residues in inter-protein interaction complexes in close spatial proximity (< 7Å) were considered as interface residues and other residues (inter-residue distances > 7 Å) were considered as non-interface residues.
2.4. Studying intra-protein co-evolution using the Co-Var methodology
A set of 252 conserved domain database (CDD) protein family alignments [50] with at least 80 sequences in each alignment were collected to study intra-molecular co-evolution (supplementary data 3 Table S2). Intra-molecular co-evolution in these protein families was studied with the help of Co-Var, CAPS [21], MI [51]and PSICOV [12] respectively. Methods were run considering default optimal parameters and the inter-residue distances among intra-protein co-evolved pairs predicted utilizing these programs were determined for comparison.
2.5. Studying co-evolution in intercellular protein–protein interaction complexes
In this study, inter-protein co-evolution has been studied in some intercellular protein interaction complexes having available mutation data. Complexes between proteins like colony stimulating factor 3 receptor (CSF3R), colony stimulating factor 3 (CSF3); transforming growth factor alpha (TGFA), epidermal growth factor receptor (EGFR); fibroblast growth factor receptor 1 (FGFR1), fibroblast growth factor 1 (FGF1); transforming growth factor beta receptor 2 (TGFBR2), transforming growth factor beta 3 (TGFB3); fibroblast growth factor receptor 2 (FGFR2), fibroblast growth factor 10 (FGF10) and fibroblast growth factor receptor 2 (FGFR2), fibroblast growth factor 1 (FGF1) were considered. The respective sequences and structures were obtained from the UniProt and PDB databases [52], [53], [54], [55]. Orthologs in each family were determined with the help of DELTA-BLAST [46] and taxonomy matched non-redundant sequences having E-value <= 1E-04, query coverage >= 70%, sequence identity >= 45% were utilized for preparing MSAs in MAFFT [47]. Co-evolving residue positions among the representative sequences in each sequence family involved in the interaction complex were determined with the help of Co-Var methodology considering z-score <= −1 as the selection threshold. Subsequently, co-evolved residue pair positions were mapped onto the corresponding 3D structure of the reference sequence to determine the inter-residue distances for a distance distribution analysis. The degree of each residue position among the co-evolved pairs was determined by analyzing a network representing residue positions as nodes and co-evolutionary pairings between positions as edges. Based on whether a residue position had a degree higher than the median of the degree distribution, high degree co-evolved positions were selected for further analysis.
2.6. Investigating the functional and physiological relevance of high degree co-evolutionary pairings
High degree co-evolved positions identified in inter-protein interactions were determined and represented on a Circosplot [49] for easy interpretation. The functional relevance of the predicted co-evolutionary pairings was analyzed by domain analysis and mutation mapping. Details regarding the domains in each protein were obtained by querying the Pfam database [56] and in-house perl programs were utilized to determine whether the residue positions involved in co-evolutionary pairings occurred within the functional domain regions of the interacting proteins. Additionally, mutation data from the COSMIC database [57] has been considered to map whether residue positions important from the standpoint of inter-molecular co-evolution exhibit frequent substitution mutations in disease conditions such as cancer. As a result of the mutation, it is plausible that the interaction between these proteins is perhaps compromised in conditions such as cancer. Amino acid pairing frequencies among the overall predicted co-evolved positions in the native reference sequences in each case were compared to the ones obtained based on the assumption that the sequences exhibit the substitution mutations.
2.7. Co-Var web server for predicting intra- and inter molecular co-evolutionary pairings
To provide a wider scope to the Co-Var methodology, we have developed a web server version of the method which is freely accessible. Utilizing a set of homologous sequences or alignment(s) of proteins as input(s) the Co-Var methodology may be utilized for studying intra-protein or inter-protein co-evolution in our Co-Var web server available via http://www.hpppi.iicb.res.in/ishi/covar/index.html. The front end of the server is HTML, PHP and java based while a perl based implementation of the Co-Var methodology works on the backend of the server to predict reference sequence (first sequence in the alignment) mapped co-evolved residue positions. Further, based on an uploaded reference structure inter-residue distances between the co-evolutionary pairings and structural mapping of pairings can be obtained. Co-evolutionary pairings in close structural proximity can be visualized in a viewer [58]. Additional modules are available for functional interpretation of the inter-protein co-evolutionary pairings in terms of their frequency of occurrence among predicted co-evolved positions (high degree co-evolved positions). Moreover, the list of reference sequence, structure mapped, and high degree co-evolutionary pairings along with reference sequence and structure mapped residue identities can be downloaded from the mailed result link.
3. Results
3.1. Studying co-evolution in protein–protein interaction complexes utilizing different analysis methods
In order to explore whether co-evolutionary connections can occur between residues in spatially proximal as well as distant regions in inter-protein interaction complexes, interacting proteins which are likely to co-evolve have been analyzed herein. The positive and negative set of complexes were studied with the help of inter-protein co-evolution analysis programs (MirrorTree [45], CAPS [37], [21] and EV-complex [33]). MirrorTree method provides a correlation coefficient as an estimation of the likelihood of co-evolution between two protein families/alignments. Initially, this measure has been utilized to verify the quality of the dataset (generated alignments) considered in this study. It has been previously shown that higher the correlation coefficient value, more likely it is that the proteins are co-evolving [59]. For our positive dataset the median of the distribution for the MirrorTree correlation co-efficient was higher than 0.8 indicating that these complexes are more likely to co-evolve than the negative set of complexes that had the median of correlation co-efficient distribution lower than 0.8 (p-value <= 0.0001) (Fig. 3A). Subsequently, analysis of inter-residue distances among the predicted co-evolved positions in interacting proteins suggested that a large percentage (65–75%) of co-evolved pairs did not lie in close spatial proximity (< 10 Å) in the results obtained from CAPS and EV-complex (Fig. 3B, 3C).
Fig. 3.
Analysis of inter-protein co-evolutionary pairings from a structural perspective. Co-evolved residue positions were mapped onto a reference structure and inter-residue distances were calculated to analyze whether the residues were in close spatial proximity (A) MirrorTree based prediction of co-evolving and non-co-evolving proteins considering the positive and negative dataset (B) Inter-residue distance distribution for EVcomplex predicted co-evolved pairs (C) Distance distribution analysis for CAPS based prediction of co-evolved pairs in interacting complexes (D) Distribution of ‘percentage of co-evolved pairs’ predicted by Co-Var in specific inter-residue distance distribution bins.
An evolutionary approach based on information theory such as Co-Var can be utilized to identify inter-dependent protein residue positions that are crucial for conservation of an interaction between two proteins. A significant fraction of residue pair positions were identified as co-evolving based on the Co-Var methodology in each of the interacting protein complexes considered herein (Supplementary data 2 Table S1). Further, in order to evaluate the efficacy of the Co-Var method in studying inter-protein co-evolution, ‘percentage of co-evolved pairs’ was determined among the interacting (positive set [50complexes]) and non-interacting proteins (negative set [50complexes]). This parameter is likely to be higher for interacting complexes which are likely to co-evolve rather than for non-interacting proteins which are less likely to co-evolve. In general, interacting complexes had a higher percentage of co-evolving pairs than the non-interacting complexes as suggested by inter-protein co-evolution analysis programs such as CAPS and EV-complex (Fig. 4A, 4B). The Co-Var methodology identified that interacting (positive) complexes had ‘percentage of co-evolved pairs’ index in the range of 6–10% while the non-interacting (negative) complexes had much lower values which occurred in the range of 0–2% and as such this index allowed the segregation of the two set of complexes substantially (p-value <= 0.0001) (Fig. 4C, Table 1). However, while the distributions of the measures calculated for the positive and negative set of complexes are segregated based on the results from EV-complex and CAPS as well, the significance obtained is lower than that with Co-Var (Fig. 4A–4C, Table 1). A closer look at the figures also revealed that overlap of ‘percentage of co-evolved pairs’ data points between the positive and negative set are far lower in Co-Var than that in EV-complex or CAPS (Fig. 4A-4C). Therefore, this analysis indicated that the Co-Var methodology performs better in determining whether a protein–protein interaction complex is likely to co-evolve and can identify actual residue pair positions exhibiting inter-dependent changes. Utilizing the Co-Var co-evolution analysis data for interacting proteins, we again observed the trend that a large percentage of co-evolved positions (nearly 70%) did not occur in close spatial proximity (<10 Å) (Fig. 3D). This trend was consistently observed in the co-evolving pairs determined among interacting proteins with the help of multiple inter-protein co-evolution analysis methodologies (Fig. 3B–3D).
Fig. 4.
Comparison Co-Var methodology with other inter-protein co-evolution analysis programs. A set of interacting proteins which are likely to co-evolve (Positive) and a set of non-interacting proteins (Negative) that are less likely to co-evolve were studied with the help of multiple programs available to study inter-protein co-evolution. (A) Inter-protein co-evolution analysis in CAPS considering the positive and negative dataset (B) Evolutionary coupling analysis (EVcomplex) to determine co-evolution in positive and negative dataset. (C) Analysis of positive and negative dataset with the help of Co-Var (D) Percentage of predicted co-evolved residue pairs that occur at the interface and percentage of interacting pairs that were found to co-evolve among the positive set of complexes analyzed utilizing Co-Var.
Table 1.
Analysis of inter-protein co-evolution utilizing Co-Var and other existing methodologies. Statistics for student’s t-test performed utilizing co-evolution parameters obtained for interacting (positive) and non-interacting (negative) proteins in Co-Var, MirrorTree, CAPS and EV-complex has been outlined here.
| Co-Var | MirrorTree correlation coefficient | CAPS | EV-complex | |||||
|---|---|---|---|---|---|---|---|---|
| Positive (n = 50) | Negative (n = 50) | Positive (n = 50) | Negative (n = 50) | Positive (n = 50) | Negative (n = 50) | Positive (n = 50) | Negative (n = 50) | |
| Mean | 7.73 | 1.26 | 0.84 | 0.42 | 0.01 | 0.04 | 7.275 | 0.729 |
| Standard deviation | 2.71 | 0.75 | 0.12 | 0.34 | 0.019 | 0.1 | 6.034 | 2.5 |
| T-test statistics | t = 16.2703 df = 98 | t = 8.2369 df = 98 | t = 2.0840 df = 98 | t = 6.8875 df = 96 | ||||
| P-value | <= 0.0001 | <= 0.0001 | 0.0398 | <=0.0001 | ||||
Moreover, in general only about 13.6% (average) of the interface pairs (residues having non-covalent interactions as determined in PISA and residues within 7 Å) exhibited a tendency to co-evolve in these protein–protein interaction complexes (Fig. 4D, Table S1).
In addition, the predicted co-evolved pairs from EV-complex and Co-Var were compared (We did not include CAPS data for comparison because it had predicted very few or zero co-evolved positions for most of the complexes). Average PC and IC values for Co-Var and EV-Complex [33] were 13.18, 0.63% and 17.96, 0.70%, respectively. Thus, similar results were obtained from Co-Var and EV-complex (Tables S3). Although these programs have completely different analysis approaches, both EV-complex and Co-Var predicted common co-evolved pairs (Tables S3). The differences may be due to differing sensitivities to background conservation, as observed in previous studies [60], [19]. Among the commonly predicted co-evolved pairs, a substantial percentage had inter-residue distances higher than 10 Å (Supplementary data 1 Fig. S1). Further, Co-Var and EV-complex showed similar average IC and PC values, which suggested that non-interface pairs were predicted as co-evolving (Fig. 4D, S1, Supplementary data 4 Table S3). Based on these observations, we believe that non-interface positions also tend to co-evolve.
Table 3.
High degree co-evolutionary pairings in inter-protein interaction complexes. Considering residue positions as nodes and co-evolutionary pairings as edges, residue positions that had many co-evolutionary connections with multiple other residues have been determined. Percentages of co-evolved residue positions that had multiple co-evolutionary connections along with positions that could be frequently prone to substitution mutations in cancer are reported.
| Protein A | Protein B | aCP in FD (%) | bHD CP with mutations (%) | cResidues forming HD CP (Protein A) | dResidues forming HD CP (Protein B) | |
|---|---|---|---|---|---|---|
| Case 1 | CSF3 (P09919) | CSF3R (Q99062) | 78.74 | 45.36 | 58 | 68 |
| Case 2 | EGFR (P00533) | TGFA (P01135) | 32.74 | 62.78 | 91 | 9 |
| Case 3 | FGF1 (P05230) | FGFR1 (P11362) | 79.42 | 53.59 | 34 | 23 |
| Case 4 | TGFB3 (P10600) | TGFBR2 (P37173) | 84.43 | 55.71 | 53 | 45 |
| Case 5 | FGF10 (O15520) | FGFR2 (P21802) | 78.68 | 52.83 | 34 | 31 |
| Case 6 | FGF1 (P05230) | FGFR2 (P21802) | 76.68 | 59.40 | 28 | 23 |
CP in FD (%): Percentage of co-evolutionary pairings among residues in functional domains.
HD CP with mutations (%): Percentage of high degree co-evolutionary pairings with mutations.
Residues forming HD CP (Protein A): Number of residues involved in high degree co-evolutionary pairings in representative protein from family A.
Residues forming HD CP (Protein B): Number of residues involved in high degree co-evolutionary pairings in representative protein from family B.
3.2. Intra-protein and inter-protein co-evolutionary connections may occur among residue pairs that are not in close spatial proximity
Studies on intra-molecular co-evolution in proteins have suggested that residues in proximity are likely to be highly co-evolving; alternately distant sites having a functional dependence are likely to co-evolve as well [21], [18]. Thus, intra-molecular co-evolution was also studied in a set of proteins utilizing Co-Var, CAPS, MI and PSICOV to study the pattern of pair-wise residue distances among the predicted intra-protein co-evolved positions. A similar trend of co-evolving positions occurring in close spatial proximity and in distal regions was observed when we determined co-evolving residues within proteins (252 CDD) [50] protein families) (Fig. 5). Additionally, this analysis suggested that Co-Var may be utilized to study intra-protein co-evolution as well since it predicts a higher percentage of co-evolved positions occurring in proximity in comparison to the previously established methodologies. Therefore, considering pair-wise residue distances among intra-protein co-evolved pairs, we observed that a higher proportion of pairs occurred in proximity in comparison to inter-protein co-evolving positions (Fig. 3D, Fig. 5A). Inter-residue distances among the co-evolved pairs in protein–protein complexes had a significant fraction of positions that did not lie in close spatial proximity as indicated by the observed range of IC values as well (Fig. 4D).
Fig. 5.
Studying intra-protein co-evolution utilizing Co-Var. Distance distribution analysis of co-evolved positions predicted within proteins was considered to evaluate the performance of Co-Var against existing methods for studying intra-protein co-evolution. (A) Inter-residue distance among predicted co-evolved pairs within proteins based on Co-Var. (B) Distance distribution analysis of intra-protein co-evolved pairs according to CAPS. (C) Inter-residue distance among predicted co-evolved pairs within proteins utilizing mutual information. (D) Distance distribution analysis considering intra-protein co-evolving pairs determined in PsiCov.
3.3. Studying co-evolution in hetero-dimeric protein complexes involved in intercellular interactions
Interestingly, with the help of multiple co-evolutionary analysis strategies, we observed a trend that co-evolved positions can occur in interface and non-interface regions. Since, Co-Var performed relatively better than the other inter-protein co-evolution analysis programs considered in this study (Fig. 4), it was utilized to determine co-evolutionary pairings in a set of hetero-dimeric protein complexes (Table 2). This analysis was performed to examine the relevance of the co-evolutionary connections between the non-interface residues. Predicted co-evolved positions in these selected protein–protein interaction complex case studies also suggested that a small fraction of co-evolved positions occur at the interface (about 0.1–0.4% of the total co-evolved pairs were found at the interface) (Table 2). Additionally, in general about 1.5–16% (mean = 5.56%) of the interface pairs were found to co-evolve in the complexes considered and this is in correlation with the observation that transient interfaces exhibit a low degree of co-evolution (Mintseris et al., 2005).
Table 2.
Co-evolution analysis in hetero-dimeric protein complexes involved in inter-cellular interactions. Co-evolutionary pairings were identified utilizing Co-Var and a z-score threshold of z <= −1 to study co-evolution in inter-cellular protein interaction complexes.
| Reference PDB structure | Reference sequence (Family A) | Reference sequence (Family B) | aPC | bIC | |
|---|---|---|---|---|---|
| Case 1 | 2D9Q | CSF3 (P09919) | CSF3R (Q99062) | 16.883 | 0.184 |
| Case 2 | 1MOX | EGFR (P00533) | TGFA (P01135) | 2.203 | 0.161 |
| Case 3 | 1EVT | FGF1 (P05230) | FGFR1 (P11362) | 3.185 | 0.15 |
| Case 4 | 1KTZ | TGFB3 (P10600) | TGFBR2 (P37173) | 5.66 | 0.053 |
| Case 5 | 1NUN | FGF10 (O15520) | FGFR2 (P21802) | 1.429 | 0.125 |
| Case 6 | 1DJS | FGF1 (P05230) | FGFR2 (P21802) | 4 | 0.374 |
PC: Percentage of interface pairs that are predicted to be co-evolved.
IC: Percentage of co-evolved pairs that occur at the interface.
In order to study the probable significance of co-evolutionary pairings which are not occurring in close spatial proximity, we have studied co-evolution patterns in certain receptor-ligand protein hetero-dimers that are known to interact aberrantly during cancer metastasis. A large fraction of co-evolutionary connections between proteins involved in intercellular interactions did not occur in close spatial proximity (Table 2, Tables S4). However, they were predominantly found to occur only within functional domains of the proteins involved in the interaction (Fig. 6). Further, in most of the protein interaction complex case studies analyzed herein about 70–80% of the positions involved in co-evolutionary pairings occur within functional domain regions of the interacting proteins (Table 3). This observation suggests that these residue positions could be biologically relevant for functional integrity of the complex.
Table 4.
Intra-protein co-evolving positions in proteins constituting a complex are also predicted as high degree inter-protein co-evolved positions. Inter-protein co-evolving residue positions that have large number of co-evolutionary connections (high degree) between proteins are likely to be important for intra-molecular co-evolution as well.
| Reference sequence (Protein family A) | Reference sequence (Protein family B) | aIntra-protein CP (A) | bIntra-protein CP (B) | cInter-protein CP (A) | dInter-protein CP (B) | eInter-protein and intra-protein CP (A) | fInter-protein and intra-protein CP (B) | |
|---|---|---|---|---|---|---|---|---|
| Case 1 | CSF3 (P09919) | CSF3R (Q99062) | 79 | 272 | 68 | 187 | 58 | 67 |
| Case 2 | EGFR (P00533) | TGFA (P01135) | 120 | 40 | 251 | 23 | 37 | 13 |
| Case 3 | FGF1 (P05230) | FGFR1 (P11362) | 56 | 166 | 36 | 143 | 34 | 23 |
| Case 4 | TGFB3 (P10600) | TGFBR2 (P37173) | 116 | 161 | 104 | 95 | 53 | 45 |
| Case 5 | FGF10 (O15520) | FGFR2 (P21802) | 85 | 107 | 42 | 88 | 34 | 28 |
| Case 6 | FGF1 (P05230) | FGFR2 (P21802) | 55 | 171 | 33 | 101 | 28 | 21 |
Intra-protein CP (A): Number of residue positions involved in intra-protein co-evolutionary pairings in reference protein of family A.
Intra-protein CP (B): Number of residue positions involved in intra-protein co-evolutionary pairings in reference protein of family B.
Inter-protein CP (A): Number of residue positions involved in inter-protein co-evolutionary pairings in reference protein of family A.
Inter-protein CP (B): Number of residue positions involved in inter-protein co-evolutionary pairings in reference protein of family B.
Inter-protein and intra-protein CP (A): Number of residue positions important for inter-protein (high degree) and intra-protein co-evolution (Protein A).
Inter-protein and intra-protein CP (B): Number of residue positions important for inter-protein (high degree) and intra-protein co-evolution (Protein B).
Fig. 6.
Co-evolving residue positions in inter-protein interactions lie predominantly within functional domain regions within each protein of the complex. Z-scores corresponding to residue positions involved in predicted co-evolutionary pairings (Z-score <=- 1) have been plotted (A) Predicted co-evolutionary pairing positions observed between CSF3 and CSF3R. (B) Predicted co-evolutionary pairings positions in TGFA and EGFR complex (C) Predicted co-evolutionary pairing positions occurring between FGF1 and FGFR1 (D) Predicted co-evolutionary pairing positions in TGFBR2 and TGFB3 complex (E) Predicted co-evolutionary pairing positions observed between FGF10 and FGFR2 (F) Predicted co-evolutionary pairing positions in FGF1 and FGFR2 complex.
3.4. High degree co-evolutionary pairings occur at interface and non-interface regions in inter-protein interaction complexes
Structural mapping of the co-evolutionary pairings indicated that several connections can exist between the residues in interacting proteins giving rise to some hub residues with a large number of connections. In order to visualize these connections between the residues (nodes), a chord diagram representing the co-evolutionary connections as arcs between the nodes (residue positions) as fragments on a circle can be considered. In this respect, considering the TGF-A and EGFR interaction complex, co-evolutionary pairings were obtained between 23 out of total 161 residue positions in TGF-A and 251 out of 1210 residue positions in EGFR, respectively. Further, most co-evolutionary pairings were obtained between certain TGF-A (9) and EGFR (91) residues resulting in a tendency for residues to have large number of co-evolutionary connections in the interacting protein partner (Fig. 7A). Moreover, co-evolutionary connections among high degree residue positions occur at the interface as well as non-interface regions (Fig. 7B).
Fig. 7.
High degree co-evolved positions in inter-cellular protein interaction complex involving TGF-A and EGFR. Residues predicted as co-evolved in inter-protein interaction complexes tend to have multiple co-evolutionary connections or pairings among them. (A) Co-evolving residues in TGF-A and EGFR that tend to have multiple co-evolutionary connections (High degree co-evolved positions) or pairings among them are shown here. (B) High degree co-evolved positions mapped onto the reference structure (PDB ID: 1MOX) lie in spatially proximal and distal regions. In the structural representation of co-evolved positions EGFR is depicted in light green while TGFA is depicted is light blue. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Additionally, considering the CSF3-CSF3R complex, co-evolutionary pairings were obtained between 68 out of total 207 residue positions in CSF3 and 187 out of 836 residue positions in CSF3R, respectively. However, most co-evolutionary pairings were obtained between certain CSF3 (58) and CSF3R (68) residues only. Here, we observed that certain residue positions exhibit a tendency to have large number of co-evolutionary connections with positions in the interacting protein partner. Thus, these co-evolutionary pairings which exist between residues in CSF3 and CSF3R comprise of 58 residue positions having inter-connections with 68 positions in the interacting protein partner resulting in a large number of co-evolutionary connections among them (Supplementary Fig. S2A). Co-evolutionary connections occurring between high degree co-evolved positions in this protein pair are present at the interface as well as non-interface regions and these could be important for the interaction between these proteins (Supplementary Fig. S2B).
Further, in the TGFB3 and TGFBR2 interaction complex, co-evolutionary pairings were obtained between 104 out of total 412 residue positions in TGFB3, and 95 out of 567 residue positions in TGFBR2 respectively. Further, most co-evolutionary pairings were obtained between certain TGFB3 (53) and TGFBR2 (45) residues again exhibiting this tendency of few residues in one binding partner to have large number of co-evolutionary connections with certain residues in the interacting protein partner (Supplementary Fig. S3A). Additionally, co-evolutionary connections were noted among high degree residue positions at the interface and non-interface regions in this protein pair as well (Supplementary Fig. S3B). Similarly, the co-evolutionary pairings obtained in the other inter-cellular interaction complexes also exhibit this tendency wherein certain residue positions exhibit a tendency to have larger number of co-evolutionary connections with positions in the interacting protein partner (Supplementary Figs. S4, S5, S6). High degree residue positions involved in co-evolutionary pairings occur in interface (spatially proximal < 7 Å) and non-interface (spatially distal > 7 Å) regions have been shown on the representative structures (Fig. 8). It is possible that such threshold selected co-evolutionary pairings with significant scores and high degree are likely to be crucial for functional protein interactions.
Fig. 8.
High degree co-evolved positions observed in interface and non-interface regions. Co-evolutionary pairings may include residue positions that have many co-evolutionary connections or pairings among them. Such high degree co-evolved positions when mapped onto reference structures were found to occur in interface and non-interface regions as represented here. High degree co-evolved positions in (A) CSF3 and CSF3R (B) TGFA and EGFR (C) FGF1 and FGFR1 (D) TGFBR2 and TGFB3 (E) FGF10 and FGFR2 (F) FGF1 and FGFR2 complex, respectively are shown in ball and stick models.
3.5. Identification of disease associated changes in intercellular protein–protein interaction complexes involved in cancer metastasis
It would be interesting to ascertain whether the high degree co-evolutionary pairings have a biological significance. For this purpose, we determined whether the residue positions occur within the functional domain regions in each protein, are important in intra-molecular co-evolution or could be frequently prone to mis-sense substitutions. The Co-Var methodology was utilized to study co-variation within each protein involved in the inter-cellular protein interaction complexes considered herein. This analysis was performed to determine whether the high degree co-evolved residue positions identified in inter-molecular co-evolution could additionally have crucial roles within a protein based on intra-molecular co-evolution as well. Intra-molecular co-evolution analysis of the complex constituent proteins demonstrated that positions important for protein stability or function are also involved in forming extensive high degree co-evolutionary pairings in inter-protein interaction complexes. Therefore, residue positions important for intra-protein stability or function may additionally influence inter-protein co-evolution interactions as well (Table 4). However, certain positions involved in inter-protein co-evolutionary pairings were only important during inter-protein co-evolution.
Generally, residues that exhibit mis-sense substitution mutation could be associated with a protein’s function. Similarly, interdependent co-evolving residues exhibiting substitution mutations could also be functionally important in inter-protein interactions. In particular, co-evolutionary pairings in complexes which exhibit aberrant protein functionality have been studied to explore this possibility. An interesting observation regarding the high degree co-evolved positions or the residue positions with large number of co-evolutionary connections was that large fractions among them were prone to substitution mutations prevalent in cancer (Table 3). Thus, high degree co-evolutionary pairings among residues that are frequently exhibit substitution mutations in diseases such as cancer could potentially be important for conservation of a functional interaction between the proteins. Moreover, we have also studied the residue pairing propensity (frequently observed residue pairs) among the predicted high degree co-evolutionary pairings. We determined that the residue pairing propensity varied substantially in mutated protein complexes wherein pairs containing amino acids such as glycine, proline, aspartate, glutamate, tryptophan, tyrosine, histidine, and glutamine are more frequent (Fig. 9). Such substitutions or alterations in pairing propensity at crucial positions particularly at interface positions that tend to co-evolve are likely to have deleterious functional characteristics in the absence of coordinated compensatory changes. A similar trend is observed in most of the intercellular protein interaction complexes considered herein where the residue pairing propensity among the co-evolved positions is likely to be altered because of disease-causing mutations (Table 3, Fig. 9). Based on these observations, it can be postulated that co-evolved residue positions could be frequently mutated in diseases (for instance, cancer) and as such alterations at these residue positions may not always have compensatory changes which could result in a perturbed interaction between these proteins. Therefore, with the help of the Co-Var methodology one can predict high degree co-evolutionary pairings in interacting proteins which may or may not be in proximity but are likely to be functionally relevant or important for maintaining an inter-protein interaction. Further, absence of coordinated changes at such interface and non-interface co-evolving residue positions may lead to disruptions in protein–protein interactions and such alterations could be disease associated.
Fig. 9.
Predicted high degree co-evolved positions may be functionally relevant in protein–protein interactions. Residue pairing propensity at the high degree co-evolved positions in reference protein sequences of the complex and the altered pairing propensity based on the observed substitution mutations have been compared. High degree co-evolved positions are frequently prone to substitution mutations and altered residue pairing propensity at these positions have been highlighted with *. (A) Pairing propensity in native CSF3 and CSF3R complex (B) Altered pairing propensity in CSF3 and CSF3R complex (C) Pairing propensity in native TGFA and EGFR complex (D) Altered pairing propensity in mutated TGFA and EGFR complex (E) Pairing propensity in native FGF1 and FGFR1 complex (F) Altered pairing propensity in mutated FGF1 and FGFR1 complex (G) Pairing propensity in native TGFBR2 and TGFB3 complex (H) Altered pairing propensity in mutated TGFBR2 and TGFB3 complex (I) Pairing propensity in native FGF10 and FGFR2 complex (J) Altered pairing propensity in mutated FGF10 and FGFR2 complex (K) Pairing propensity in native FGF1 and FGFR2 complex (L) Altered pairing propensity in mutated FGF1 and FGFR2 complex.
3.6. Co-Var web server for studying intra-protein and inter-molecular co-evolution
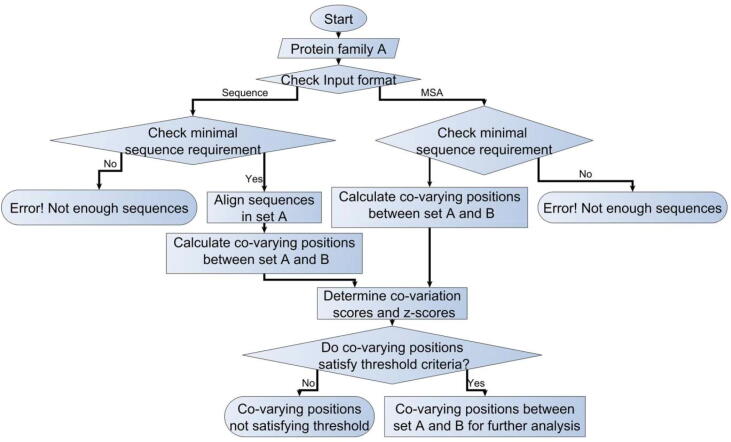
A web server for analyzing intra-protein and inter-molecular co-evolution is available online at http://www.hpppi.iicb.res.in/ishi/covar/index.html (Fig. 10A). During the inter-protein co-evolution analysis, co-evolutionary pairings are determined based on the Co-Var methodology and reference sequence mapped co-evolving positions are reported with the help of a surface plot representation. The Co-Var score and z-score for threshold selected co-evolutionary pairings are depicted (Fig. 10B). Moreover, residue identities based on the reference sequence and structure are mentioned in the list of co-evolved positions. High degree co-evolved positions and/or co-evolved positions in spatial proximity are displayed on the reference structure provided and the list of co-evolving positions in close spatial proximity may be downloaded (Fig. 10B). Further, a distance distribution plot of the inter-residue distances among co-evolved position pairs provided can be utilized to get an idea about whether co-evolutionary pairings are occurring in proximity or among spatially distant residue positions. Additionally, high degree co-evolved positions that are likely to be important for maintaining inter-protein functional interaction are also determined, and the same are provided as lists. The results of the analysis are mailed to the e-mail address provided and are easily available for download.
Fig. 10.
Co-Var web server to study intra-protein and inter-molecular co-evolution. (A) Co-Var web-server user interface (B) Snap-shot of inter-protein co-evolution analysis results provided by Co-Var web-server.
4. Discussion
Molecular co-evolution refers to a phenomenon where a change in one locus is likely to affect the selection pressure at another locus and a reciprocal change may occur reflecting a direct evolutionary interaction. Such an evolutionary interaction could be occurring between sites within a single protein referred to as intra-protein co-evolution or between different proteins in which case it is referred to as inter-protein co-evolution [31]. In this study, we have developed the Co-Var methodology which utilizes mutual information and Bhattacharyya co-efficient to study intra-protein and inter-protein co-evolution. Multiple methodologies have previously been developed to study intra-protein (MI, CAPS, and PSICOV) and inter-protein (CAPS, EV-complex) co-evolution [21], [51], [12], [33]. However, our approach has some advantages such as non-dependence on identical alignment lengths and a reduced influence of phylogenetic relationships of the organisms/species represented in the alignment. This is because supplied sequences in the alignment are randomly selected for the analysis and alignment shuffling is also performed. Further, the Co-Var methodology described here has also been implemented into an easy to use more generic Co-Var web server platform. Herein, probable co-evolutionary connections within proteins and across biomolecules or complexes, such as protein–protein, can be estimated and their structural and functional relevance can be judged. Moreover, the inter-protein co-evolution analysis platform has been extensively validated and its application has been exemplified with the help of some inter-cellular protein interaction complexes. Detailed analysis has been performed to identify whether co-evolutionary pairings occur at the interaction interface or other regions important for complex recognition, formation, or functionality.
Studies pertaining to intra-molecular co-evolution have identified that coevolving positions occur in two different categories. The first set comprises of positions that co-evolve with only one or two other positions and often exhibit direct amino acid side-chain interactions with their coevolving partner in proximity. However, the second set includes positions that co-evolve with many other positions which are predominantly located in regions critical for protein function, for instance active sites or regions involved in intermolecular interactions and recognition [20], [18]. In a similar manner the Co-Var methodology can be utilized to study intra-molecular co-evolution in proteins. For intra-molecular co-evolution analysis using the CDD protein families we found that in general it predicts a higher fraction of co-evolved residues in proximity than the other programs that had been considered during this analysis.
Co-evolution is also evident in biological systems where the interaction patterns must be maintained while the interactions continue to evolve and acquire new functions and/or avoid crosstalk with other available systems. This scenario is prevalent in signalling cascades, where a rapid divergence may occur to avoid interference with the original system. However, such a change is generally compensated by the interacting partners to maintain a functional cascade [61]. Inter-molecular co-evolution studies in proteins have shown that residues in close spatial proximity at the interaction interface generally exhibit a higher tendency to co-evolve than other residue pairs predicted as co-evolving which are spatially separated [30], [38]. Here, in our analysis, we could find that a certain fraction of co-evolving residue pairs were predicted in spatially separated positions via multiple inter-protein co-evolution analysis programs that we have utilized. Further, these co-evolutionary pairings that occur at interface or non-interface regions occur among residues present in functional domains or residues with roles in intra-protein co-evolution in the individual constituent proteins of the complex. Moreover, with the help of inter-protein co-evolution analysis of intercellular complexes involved in cancer metastasis we have determined that certain receptor positions share many co-evolutionary pairing connections with multiple ligand positions and vice versa. Such positions termed as high degree co-evolutionary pairings have been found to be frequently prone to mis-sense substitution mutations in cancer and as such absence of coordinated changes at these positions may contribute to altered interaction complexities. Therefore, the Co-Var methodology allows one to predict high degree co-evolving residue pair positions; alterations at which could be functionally detrimental for a protein–protein interaction to occur. Moreover, it has been identified that co-evolutionary pairings crucial for functional interactions in inter-protein complexes may occur in close spatial proximity or at non-interface regions. Based on these analyses, we could determine that lack of coordinated changes at co-varying residue positions could be a likely contributing factor to the altered functionality of complexes involved in processes such as cancer metastasis. In this manner, one can ascertain co-evolutionary pairings that are likely to be crucial for functional interactions between proteins which when altered could be disease associated. Thus, the information theory-based Co-Var measure may be utilized to study interacting proteins that co-evolve and to determine co-evolutionary pairings among residues that could be structurally or functionally relevant for inter-protein interactions.
5. Data availability
The data pertaining to the conclusions in this article are available in the article and in its online supplementary material. Data utilized for arriving at the conclusions presented in the work may be downloaded from http://www.hpppi.iicb.res.in/ishi/covar/about.html or http://www.hpppi.iicb.res.in/ishi/covar/download/CoVar-dat.tar.gz. It is also available on the Dryad repository (Mukherjee, Ishita; Chakrabarti, Saikat (2020), Dataset for article “Co-evolutionary landscape at the interface and non-interface regions of protein–protein interaction complexes”, Dryad, Dataset, https://doi.org/10.5061/dryad.zgmsbcc8g). A local version of the Co-Var method to determine co-evolutionary pairings in biomolecules is available for download from the Co-Var web server (http://www.hpppi.iicb.res.in/ishi/covar/about.html or http://www.hpppi.iicb.res.in/ishi/covar/download/covar-loc.zip) and/or the GitHub repository (https://github.com/Ishita2690/Co-Var).
6. Author’s contributions
Ishita Mukherjee: Data curation, Conceptualization, Methodology, Formal analysis, Software, Validation, Visualization, Investigation, Writing- Original draft preparation, Writing - review & editing. Saikat Chakrabarti: Conceptualization, Investigation, Methodology, Formal analysis, Project administration, Writing- Original draft preparation, Supervision, Reviewing and Editing, Funding acquisition, Resources.
Funding
This work was supported by the Department of Science and Technology, New Delhi, India [DST HRR fund “GAP362”]. This work was also partially funded by IICB lab reserve fund. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
CRediT authorship contribution statement
Ishita Mukherjee: Data curation, Conceptualization, Methodology, Formal analysis, Software, Validation, Visualization, Investigation, Writing- Original draft preparation, Writing - review & editing. Saikat Chakrabarti: Conceptualization, Investigation, Methodology, Formal analysis, Project administration, Writing- Original draft preparation, Supervision, Reviewing and Editing, Funding acquisition, Resources.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Authors would like to acknowledge the initial contribution of Abhijit Chakraborty in the project. We would also like to thank Sunandan Dhar for his involvement in this work during his summer project tenure. SC acknowledges CSIR-Indian Institute of Chemical Biology (IICB) for infrastructural support. IM is thankful to CSIR for her fellowship.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.06.039.
Contributor Information
Ishita Mukherjee, Email: mukishi@gmail.com, ishitamukherjee90@csiriicb.res.in.
Saikat Chakrabarti, Email: saikat@iicb.res.in.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Fitch W.M. Rate of change of concomitantly variable codons. J Mol Evol. 1971;1(1):84–96. doi: 10.1007/BF01659396. [DOI] [PubMed] [Google Scholar]
- 2.Galtier N. Maximum-likelihood phylogenetic analysis under a covarion-like model. Mol Biol Evol. 2001 May;18(5):866–873. doi: 10.1093/oxfordjournals.molbev.a003868. PMID: 11319270. [DOI] [PubMed] [Google Scholar]
- 3.Pazos F., Valencia A. Protein co-evolution, co-adaptation and interactions. EMBO J. 2008;27(20):2648–2655. doi: 10.1038/emboj.2008.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakrabarti S., Panchenko A.R., Fernandez-Fuentes N. Structural and Functional Roles of Coevolved Sites in Proteins. PLoS ONE. 2010;5(1):e8591. doi: 10.1371/journal.pone.0008591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Juan D., Pazos F., Valencia A. Emerging methods in protein co-evolution. Nat Rev Genet. 2013;14(4):249–261. doi: 10.1038/nrg3414. [DOI] [PubMed] [Google Scholar]
- 6.Cocco S., Feinauer C., Figliuzzi M., Monasson R., Weigt M. Inverse statistical physics of protein sequences: a key issues review. Rep Prog Phys. 2018;81(3):032601. doi: 10.1088/1361-6633/aa9965. [DOI] [PubMed] [Google Scholar]
- 7.Figliuzzi M., Barrat-Charlaix P., Weigt M. How pairwise coevolutionary models capture the collective residue variability in proteins? Mol Biol Evol. 2018 Apr 1;35(4):1018–1027. doi: 10.1093/molbev/msy007. [DOI] [PubMed] [Google Scholar]
- 8.Morcos F., Onuchic J.N. The role of coevolutionary signatures in protein interaction dynamics, complex inference, molecular recognition, and mutational landscapes. Curr Opin Struct Biol. 2019;56:179–186. doi: 10.1016/j.sbi.2019.03.024. [DOI] [PubMed] [Google Scholar]
- 9.Marks D.S., Colwell L.J., Sheridan R., Hopf T.A., Pagnani A., Zecchina R. Protein 3D structure computed from evolutionary sequence variation. PLoS ONE. 2011;6(12):e28766. doi: 10.1371/journal.pone.0028766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morcos F., Pagnani A., Lunt B., Bertolino A., Marks D.S., Sander C. Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc Natl Acad Sci U S A. 2011;108(49):E1293–E1301. doi: 10.1073/pnas.1111471108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hopf TA, Colwell LJ, Sheridan R, Rost B, Sander C, Marks DS. Three-dimensional structures of membrane proteins from genomic sequencing. Cell. 2012;149(7):1607-21. 10.1016/j.cell.2012.04.012. Epub 2012 May 10. PMID: 22579045; PMCID: PMC3641781. [DOI] [PMC free article] [PubMed]
- 12.Jones DT, Buchan D W A., Cozzetto D & Pontil M. (2012) PSICOV: precise structural contact prediction using sparse inverse covariance estimation on large multiple sequence alignments. Bioinformatics 28, 184–190 [DOI] [PubMed]
- 13.Kamisetty H, Ovchinnikov S, Baker D. Assessing the utility of coevolution-based residue-residue contact predictions in a sequence- and structure-rich era. Proc Natl Acad Sci U S A. 2013 Sep 24;110(39):15674-9. 10.1073/pnas.1314045110. Epub 2013 Sep 5. Erratum in: Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed]
- 14.Pollock D.D., Taylor W.R., Goldman N. Coevolving protein residues: maximum likelihood identification and relationship to structure. J Mol Biol. 1999 Mar 19;287(1):187–198. doi: 10.1006/jmbi.1998.2601. PMID: 10074416. [DOI] [PubMed] [Google Scholar]
- 15.Valencia A., Pazos F. Computational methods for the prediction of protein interactions. Curr Opin Struct Biol. 2002 Jun;12(3):368–373. doi: 10.1016/s0959-440x(02)00333-0. PMID: 12127457. [DOI] [PubMed] [Google Scholar]
- 16.Choi S.S., Li W., Lahn B.T. Robust signals of coevolution of interacting residues in mammalian proteomes identified by phylogeny-aided structural analysis. Nat Genet. 2005 Dec;37(12):1367–1371. doi: 10.1038/ng1685. Epub 2005 Nov 13 PMID: 16282975. [DOI] [PubMed] [Google Scholar]
- 17.Dutheil J., Pupko T., Jean-Marie A., Galtier N. A model-based approach for detecting coevolving positions in a molecule. Mol Biol Evol. 2005;22(9):1919–1928. doi: 10.1093/molbev/msi183. [DOI] [PubMed] [Google Scholar]
- 18.Chakrabarti S., Panchenko A.R. Coevolution in defining the functional specificity. Proteins. 2009;75(1):231–240. doi: 10.1002/prot.22239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mandloi S, Chakrabarti S. (2017) Protein sites with more coevolutionary connections tend to evolve slower, while more variable protein families acquire higher coevolutionary connections. F1000Res. 2017;6:453. 10.12688/f1000research.11251.2 [DOI] [PMC free article] [PubMed]
- 20.Gloor G.B., Martin L.C., Wahl L.M., Dunn S.D. Mutual information in protein multiple sequence alignments reveals two classes of coevolving positions. Biochemistry. 2005;44(19):7156–7165. doi: 10.1021/bi050293e. [DOI] [PubMed] [Google Scholar]
- 21.Fares M.A., Travers S.A. A novel method for detecting intramolecular coevolution: adding a further dimension to selective constraints analyses. Genetics. 2006;173(1):9–23. doi: 10.1534/genetics.105.053249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aguilar D, Oliva B, Marino Buslje C. Mapping the mutual information network of enzymatic families in the protein structure to unveil functional features. PLoS One. 2012;7(7):e41430. 10.1371/journal.pone.0041430. Epub 2012 Jul 25. PMID: 22848494; PMCID: PMC3405127. [DOI] [PMC free article] [PubMed]
- 23.Cheung N.J., John Peter A.T., Kornmann B. Leri: a web-server for identifying protein functional networks from evolutionary couplings. Comput Struct Biotechnol J. 2021 doi: 10.1016/j.csbj.2021.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Süel G.M., Lockless S.W., Wall M.A., Ranganathan R. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat Struct Biol. 2003;10(1):59–69. doi: 10.1038/nsb881. [DOI] [PubMed] [Google Scholar]
- 25.Noivirt-Brik O., Horovitz A., Unger R., Onuchic J. Trade-off between positive and negative design of protein stability: From lattice models to real proteins. PLOS Comput Biol. 2009;5(12):e1000592. doi: 10.1371/journal.pcbi.1000592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacob E, Unger R, Horovitz A (2015) Codon-level information improves predictions of inter-residue contacts in proteins by correlated mutation analysis. eLife 4:e08932. [DOI] [PMC free article] [PubMed]
- 27.Wang X., Jing X., Deng Y.i., Nie Y., Xu F., Xu Y. Evolutionary coupling saturation mutagenesis: Coevolution-guided identification of distant sites influencing Bacillus naganoensis pullulanase activity. FEBS Lett. 2020;594(5):799–812. doi: 10.1002/feb2.v594.510.1002/1873-3468.13652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anishchenko I, Ovchinnikov S, Kamisetty H, Baker D. (2017) Origins of coevolution between residues distant in protein 3D structures protein structures. Proceedings of the National Academy of Sciences, 114 (34) 9122-9127; 10.1073/pnas.1702664114 [DOI] [PMC free article] [PubMed]
- 29.Salinas VH, Ranganathan R. Coevolution-based inference of amino acid interactions underlying protein function. Elife. 2018 Jul 20;7:e34300. 10.7554/eLife.34300. PMID: 30024376; PMCID: PMC6117156. [DOI] [PMC free article] [PubMed]
- 30.Mintseris J., Weng Z. Structure, function, and evolution of transient and obligate protein-protein interactions. Proc Natl Acad Sci USA. 2005;102(31):10930–10935. doi: 10.1073/pnas.0502667102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lovell S.C., Robertson D.L. An Integrated View of Molecular Coevolution in Protein-Protein Interactions. Mol Biol Evol. 2010;27(11):2567–2575. doi: 10.1093/molbev/msq144. [DOI] [PubMed] [Google Scholar]
- 32.Pazos F., Helmer-Citterich M., Ausiello G., Valencia A. Correlated mutations contain information about protein-protein interaction. J Mol Biol. 1997;271(4):511–523. doi: 10.1006/jmbi.1997.1198. [DOI] [PubMed] [Google Scholar]
- 33.Hopf T A, Schärfe C P, Rodrigues JP, Green AG, Kohlbacher O, Sander C, Bonvin AM, et al. (2014). Sequence co-evolution gives 3D contacts and structures of protein complexes. eLife, 3, e03430. 10.7554/eLife.03430 [DOI] [PMC free article] [PubMed]
- 34.Ovchinnikov S, Kamisetty H, Baker D. Robust and accurate prediction of residue-residue interactions across protein interfaces using evolutionary information. Elife. 2014 May 1;3:e02030. 10.7554/eLife.02030. [DOI] [PMC free article] [PubMed]
- 35.Marino Buslje C., Teppa E., Di Doménico T., Delfino J.M., Nielsen M., Rost B. Networks of high mutual information define the structural proximity of catalytic sites: implications for catalytic residue identification. PLoSComputBiol. 2010;6(11):e1000978. doi: 10.1371/journal.pcbi.1000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teppa E., Zea D.J., Marino-Buslje C. Protein–protein interactions leave evolutionary footprints: High molecular coevolution at the core of interfaces. Protein Sci. 2017;26(12):2438–2444. doi: 10.1002/pro.v26.1210.1002/pro.3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fares M.A., McNally D. CAPS: coevolution analysis using protein sequences. Bioinformatics. 2006;22(22):2821–2822. doi: 10.1093/bioinformatics/btl493. [DOI] [PubMed] [Google Scholar]
- 38.Andrade M., Pontes C., Treptow W. Coevolutive, evolutive and stochastic information in protein-protein interactions. Comput Struct Biotechnol J. 2019;20(17):1429–1435. doi: 10.1016/j.csbj.2019.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao M., Zhou H., Skolnick J. Insights into Disease-Associated Mutations in the Human Proteome through Protein Structural Analysis. Structure. 2015;23(7):1362–1369. doi: 10.1016/j.str.2015.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.David A., Sternberg M.J. The Contribution of Missense Mutations in Core and Rim Residues of Protein-Protein Interfaces to Human Disease. J Mol Biol. 2015;427(17):2886–2898. doi: 10.1016/j.jmb.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sowmya, G., Breen, E. J., &Ranganathan, S. (2015). Linking structural features of protein complexes and biological function. Protein science: a publication of the Protein Society, 24(9), 1486-94. [DOI] [PMC free article] [PubMed]
- 42.Mintseris J., Weng Z. Atomic contact vectors in protein-protein recognition. Proteins. 2003;53(3):629–639. doi: 10.1002/prot.v53:310.1002/prot.10432. [DOI] [PubMed] [Google Scholar]
- 43.Rodriguez-Rivas J., Marsili S., Juan D., Valencia A. Conservation of coevolving protein interfaces bridges prokaryote-eukaryote homologies in the twilight zone. Proc Natl Acad Sci USA. 2016;113(52):15018–15023. doi: 10.1073/pnas.1611861114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smialowski P., Pagel P., Wong P., Brauner B., Dunger I., Fobo G. The Negatome database: a reference set of non-interacting protein pairs. Nucleic Acids Res. 2009;38(Database issue):D540–D544. doi: 10.1093/nar/gkp1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ochoa D., Pazos F. Studying the co-evolution of protein families with the Mirrortree web server. Bioinformatics. 2010;26(10):1370–1371. doi: 10.1093/bioinformatics/btq137. [DOI] [PubMed] [Google Scholar]
- 46.Boratyn G.M., Schäffer A.A., Agarwala R., Altschul S.F., Lipman D.J., Madden T.L. Domain enhanced lookup time accelerated BLAST. Biology direct. 2012;7(1):12. doi: 10.1186/1745-6150-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on Fast Fourier transform. Nucleic Acids Res. 2002;30(14):3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dunn S.D., WahlL M., Gloor G.B. Mutual information without the influence of phylogeny or entropy dramatically improves residue contact prediction. Bioinformatics. 2008;24(3):333–340. doi: 10.1093/bioinformatics/btm604. [DOI] [PubMed] [Google Scholar]
- 49.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 50.Marchler-Bauer A., Derbyshire M.K., Gonzales N.R., Shennan L.u., FaridehChitsaz L.Y., Geer R.C. CDD: NCBI's conserved domain database. Nucleic Acids Res. 2015;43(D1):D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Korber B.T., Farber R.M., Wolpert D.H., Lapedes A.S. Covariation of mutations in the V3 loop of human immunodeficiency virus type 1 envelope protein: an information theoretic analysis. Proc. Natl. Acad. Sci. 1993;90(15):7176–7180. doi: 10.1073/pnas.90.15.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rose PW, Prlić A, Bi C, Bluhm WF, Christie CH, Dutta S, et al The RCSB Protein Data Bank: views of structural biology for basic and applied research and education. Nucleic Acids Research 2015; 43(Database issue):D345-56. [DOI] [PMC free article] [PubMed]
- 53.Tamada T., Honjo E., Maeda Y., Okamoto T., Ishibashi M., Tokunaga M. Homodimeric cross-over structure of the human granulocyte colony-stimulating factor (GCSF) receptor signaling complex. ProcNatlAcadSci U S A. 2006;103(9):3135–3140. doi: 10.1073/pnas.0511264103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Breuza L, Poux S, Estreicher A, Famiglietti M L, Magrane M, Tognolli M, et al. (2016). The UniProtKB guide to the human proteome. Database : the journal of biological databases and curation, 2016, bav120. 10.1093/database/bav120 [DOI] [PMC free article] [PubMed]
- 55.The UniProt Consortium UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47(D1):D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R. Pfam: the protein families database. Nucleic Acids Res. 2014;42(D1):D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tate, J. G., Bamford, S., Jubb, H. C., Sondka, Z., Beare, D. M., Bindal, N., et al. (2019). COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic acids research, 47(D1), D941–D947. 10.1093/nar/gky1015 [DOI] [PMC free article] [PubMed]
- 58.Rego Nicholas and Koes David (2015) 3Dmol.js: molecular visualization with WebGL Bioinformatics 31 (8): 1322-1324 10.1093/bioinformatics/btu829 [DOI] [PMC free article] [PubMed]
- 59.Herman D., Ochoa D., Juan D. Selection of organisms for the co-evolution-based study of protein interactions. BMC Bioinf. 2011;12:363. doi: 10.1186/1471-2105-12-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fodor A.A., Aldrich R.W. Influence of conservation on calculations of amino acid covariance in multiple sequence alignments. Proteins. 2004;56(2):211–221. doi: 10.1002/prot.20098. [DOI] [PubMed] [Google Scholar]
- 61.Ochoa D., Pazos F. Practical aspects of protein co-evolution.Front. Cell. Dev. Biol. 2014;2(14) doi: 10.3389/fcell.2014.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.R Core Team. (2017) R: A Language and Environment for Statistical Computing. https://www.R-project.org
- 63.Zhou J.X., Taramelli R., Pedrini E., Knijnenburg T., Huang S. Extracting Intercellular Signaling Network of Cancer Tissues using Ligand-Receptor Expression Patterns from Whole-tumor and Single-cell Transcriptomes. Sci Rep. 2017;7(1) doi: 10.1038/s41598-017-09307-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data pertaining to the conclusions in this article are available in the article and in its online supplementary material. Data utilized for arriving at the conclusions presented in the work may be downloaded from http://www.hpppi.iicb.res.in/ishi/covar/about.html or http://www.hpppi.iicb.res.in/ishi/covar/download/CoVar-dat.tar.gz. It is also available on the Dryad repository (Mukherjee, Ishita; Chakrabarti, Saikat (2020), Dataset for article “Co-evolutionary landscape at the interface and non-interface regions of protein–protein interaction complexes”, Dryad, Dataset, https://doi.org/10.5061/dryad.zgmsbcc8g). A local version of the Co-Var method to determine co-evolutionary pairings in biomolecules is available for download from the Co-Var web server (http://www.hpppi.iicb.res.in/ishi/covar/about.html or http://www.hpppi.iicb.res.in/ishi/covar/download/covar-loc.zip) and/or the GitHub repository (https://github.com/Ishita2690/Co-Var).