Figure 3.
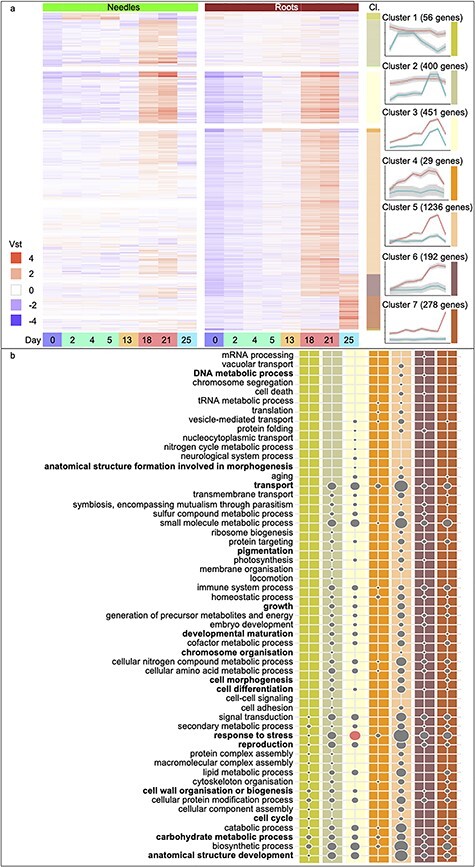
Gene Ontology Slim enrichment analysis of biological processes up-regulated in response to drought stress. Heatmaps of DEGs with expression over the eight sampling time points (Days 0, 2, 4, 5, 13, 18, 21 and 25 and colored as in Figure 1), separated by tissue, needles (green) and roots (brown). (a) Up-regulated genes of needle- (Clusters 1 and 2) and root-specific (Clusters 4–7) clusters or in common between the tissues (Cluster 3). Displayed are the Variance Stabilizing Transformation (vst) values scaled by row means. The seven highly populated clusters (Cl.) are detailed on the side, and the average trend of gene expression and a 95% CI are indicated for both tissues. Colored bars to the right of the heatmap correspond to the clusters represented in the line graphs as indicated by a colored bar to the right of each cluster line graph. (b) The lower panel of the figure lists all GOSlim biological processes categories represented in the clusters shown in (a), with columns colored by cluster. Bold text indicates categories that are significantly enriched. Circle sizes are relative to the category containing the largest number of genes; ‘response to stress’ (76 up-regulated genes). Circles in red represent significantly enriched categories (false discovery rate-adjusted P-value <0.05).
