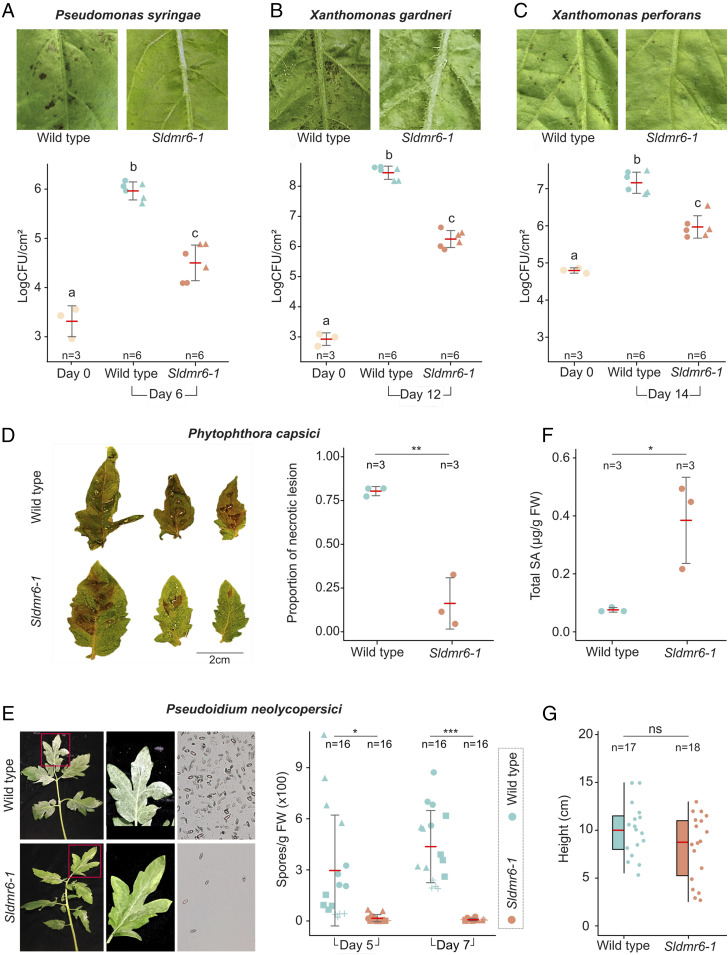
Fig. 2.
SlDMR6-1 inactivation in tomato leads to broad-spectrum disease resistance. Disease symptoms and resistance assays with the bacterial pathogens (A) P. syringae pv. tomato DC3000, (B) X. gardneri 153, and (C) X. perforans 4b, (D) the oomycete P. capsici LT1534 isolate (P = 0.00172), and (E) the fungus P. neolycopersici MF-1 isolate (PDay5 = 0.0037, PDay7 = 1.5E-06). Fungal spores in E were imaged using an epifluorescence microscope with 5× lens. All these pathogens show reduced growth or cause less disease symptoms in the Sldmr6-1 mutants. The letters indicate significant differences between the conditions as determined using a one-way ANOVA followed by a Tukey honest significant difference (HSD) test (P < 0.05). Symbols (circles, triangles, and squares) represent leaves from different plants. (F) Total SA content is shown for Fla. 8000 wild-type plants and Sldmr6-1 mutants (P value = 0.023) in response to X. gardneri infection. (G) Quantification of the heights of wild-type and Sldmr6-1 lines (P = 0.121) grown under laboratory conditions. Asterisks represent significant P values from t tests: ns, not significant, P ≥ 0.05; *P < 0.05; **P < 0.01; ***P < 0.001.