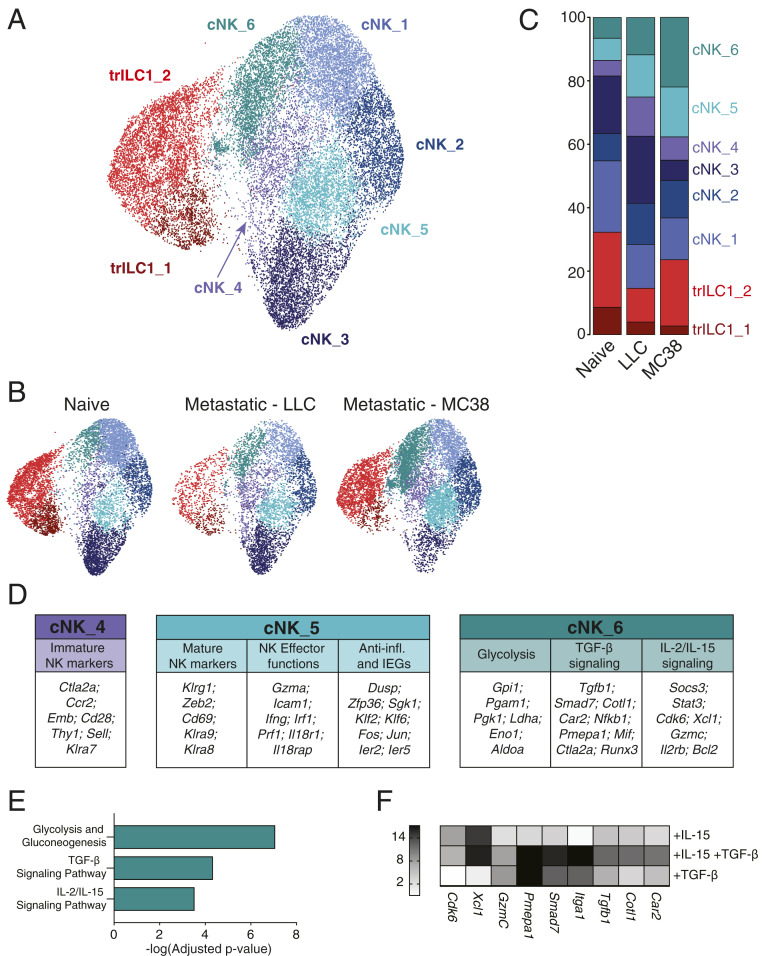
Fig. 6.
Single-cell RNA-seq reveals unique transcriptional signatures of cNKs in the hepatic metastatic niche. Single-cell RNA-seq was performed on NKp46+ cells sorted from naïve, LLC metastatic, and MC38 metastatic livers (six mice per condition pooled into one sample for droplet encapsulation and library preparation). (A–C) UMAP projections identifying the different cNK/trILC1 clusters in (A) all samples or (B) individual samples with (C) bar plots showing the relative frequency of the different clusters in each sample. (D) Top up-regulated genes in cluster cNK_4, cNK_5, and cNK_6 arranged in functional categories. (E) Pathway analysis of significantly up-regulated genes in cluster cNK_6. (F) cNKs and trILC1s were sorted from naïve livers and cultured in vitro for 48 h with 25 ng/mL mouse IL-15/IL-15R complex and/or 10 ng/mL human TGF-β1. The transcripts of cNK_6 cluster-specific genes were quantified by qPCR. The heat map shows the average fold change relative to the untreated condition. The data represent three biological and three technical replicates, and the experiment was performed three times with similar results.